Human APP Intracellular Domain Complex with Fe65-PTB2
From Proteopedia
Cleavage of the amyloid precursor protein (APP) is a crucial event in Alzheimer disease pathogenesis that creates the amyloid-beta peptide (Abeta) and liberates the carboxy-terminal APP intracellular domain (AICD) into the cytosol. [1] The study of interaction of the AICD and Fe65 protein is rather important, as Fe65 protein seems to be implicated in production of Abeta and in signalling in APP.
Contents |
Structure
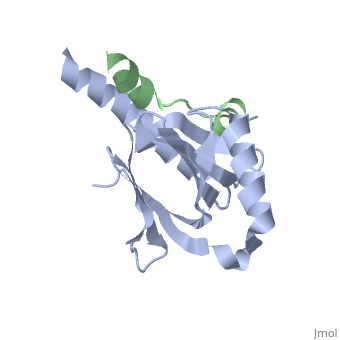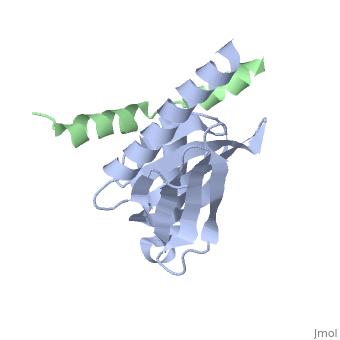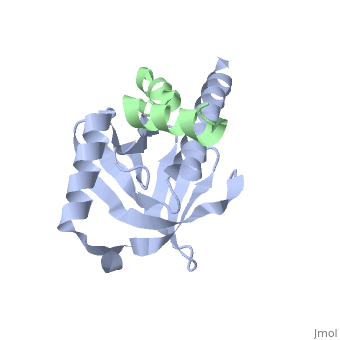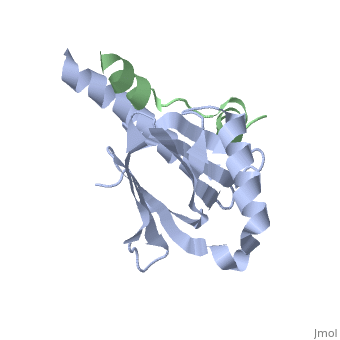
This crystal structure contains 4 chains. . Each contains 140 residues: 4 helices and 7 strands. It's a part of the protein Fe65 binded with APP intracellular domain. . Each contains 35 residues: 2 helices and 1 strand. They represent APP intracellular domain.
The APP intracellular domain is in complex with the . The interaction of the APP C terminus with the adaptor protein Fe65 mediates APP trafficking and signalling, and is thought to regulate APP processing and Abeta generation. The unique interface involves the NPxY PTB-binding motif and two alpha helices. The amino-terminal helix of the APP intracellular domain is , it's an Alzheimer disease-relevant phosphorylation site which is involved in Fe65-binding regulation. The structure together with mutational studies, isothermal titration calorimetry and nuclear magnetic resonance experiments sets the stage for understanding T(668) phosphorylation-dependent complex regulation at a molecular level.[2] Mutation at Thr-668 of APP abolished the effect of Fe65 on APP maturation. This mutation blocked the Fe65-dependent suppression of Abeta production and resulted in the release of increased levels of Abeta in the presence of Fe65 [3] .
Functions of human APP and Fe65 protein
There are suggestions that the binding of APP with Fe65 has been implicated in regulating cell motility and growth cone dynamics [4] [5]
APP is a cell surface receptor and performs physiological functions on the surface of neurons relevant to neurite growth, neuronal adhesion and axonogenesis. APP is involved in cell mobility and transcription regulation through protein-protein interactions. Can promote transcription activation through binding to APBB1-KAT5 and inhibits Notch signaling through interaction with Numb. Couples to apoptosis-inducing pathways such as those mediated by G(O) and JIP. Inhibits G(o) alpha ATPase activity By similarity. Acts as a kinesin I membrane receptor, mediating the axonal transport of beta-secretase and presenilin 1. Involved in copper homeostasis/oxidative stress through copper ion reduction. In vitro, copper-metallated APP induces neuronal death directly or is potentiated through Cu2+-mediated low-density lipoprotein oxidation. Can regulate neurite outgrowth through binding to components of the extracellular matrix such as heparin and collagen I and IV. The splice isoforms that contain the BPTI domain possess protease inhibitor activity.
Beta-amyloid peptides are lipophilic metal chelators with metal-reducing activity. Bind transient metals such as copper, zinc and iron. In vitro, can reduce Cu2+ and Fe3+ to Cu+ and Fe2+, respectively. Beta-amyloid 42 is a more effective reductant than beta-amyloid 40. Beta-amyloid peptides bind to lipoproteins and apolipoproteins E and J in the CSF and to HDL particles in plasma, inhibiting metal-catalyzed oxidation of lipoproteins. Beta-APP42 may activate mononuclear phagocytes in the brain and elicit inflammatory responses. Promotes both tau aggregation and TPK II-mediated phosphorylation. Interaction with overexpressed HADH2 leads to oxidative stress and neurotoxicity. [6] [7] [8] [9]
Fe65 is an adaptor protein localized in the nucleus. It interacts with the Alzheimer's disease amyloid precursor protein (APP), transcription factor CP2/LSF/LBP1 and the low-density lipoprotein receptor-related protein. APP functions as a cytosolic anchoring site that can prevent the gene product's nuclear translocation. It is thought to regulate transcription. Also it is observed to block cell cycle progression by downregulating thymidylate synthase expression. Multiple alternatively spliced transcript variants have been described for this gene but some of their full length sequence is not known.[10]
Fe65 plays a central role in the response to DNA damage by translocating to the nucleus and inducing apoptosis. May act by specifically recognizing and binding histone H2AX phosphorylated on 'Tyr-142' (H2AXY142ph) at double-strand breaks (DSBs), recruiting other pro-apoptosis factors such as MAPK8/JNK1.Required for histone H4 acetylation at double-strand breaks (DSBs).Its ability to specifically bind modified histones and chromatin modifying enzymes such as KAT5/TIP60, probably explains its transcription activation activity. [11]
Location
Fe65 could be located in cell membrane, cytoplasm, nucleus. In normal conditions, it mainly localizes to the cytoplasm, while a small fraction is tethered to the cell membrane via its interaction with APP. Following exposure to DNA damaging agents, it is released from cell membrane and translocates to the nucleus. Nuclear translocation is under the regulation of APP.[12]
APP is a cell surface protein that rapidly becomes internalized via clathrin-coated pits. During maturation, the immature APP (N-glycosylated in the endoplasmic reticulum) moves to the Golgi complex where complete maturation occurs (O-glycosylated and sulfated). After alpha-secretase cleavage, soluble APP is released into the extracellular space and the C-terminal is internalized to endosomes and lysosomes. Some APP accumulates in secretory transport vesicles leaving the late Golgi compartment and returns to the cell surface. Gamma-[]CTF(59) peptide is located to both the cytoplasm and nuclei of neurons. It can be translocated to the nucleus through association with APBB1 (Fe65). Beta-APP42 associates with FRPL1 at the cell surface and the complex is then rapidly internalized. APP sorts to the basolateral surface in epithelial cells. During neuronal differentiation, the Thr-743 phosphorylated form is located mainly in growth cones, moderately in neurites and sparingly in the cell body. Casein kinase phosphorylation can occur either at the cell surface or within a post-Golgi compartment [13]
APP and Alzheimer disease
In the 2006, there were 22.6 million people affected with Alzheimer disease, estimations predict by the year 2050 up to 106.8 cases worldwide. [14]
Aβ peptides are generated in neuronal secretory vesicles by proteolytic cleavage of the amyloid precursor protein (APP) by proteases, called β-secretase and γ-secretase that cleave at the N-terminus and variant C-termini of Aβ within APP, respectively, resulting in Aβ of 40 or 42 amino acids (Aβ40 and Aβ42, respectively)[15]
There is an association between Aβ protein and Alzheimer’s disease because it is the major component found in plaques that are found in Alzheimer patients. Increased levels of Aβ protein are linked to a decrease of cognitive abilities which are observed in Alzheimer patients. [16]
Besides neuritic plaques neurofibrillary tangles containing hyperphosphorylated tau protein are characteristic in the neuropathology of Alzheimer’s disease. Hyperphosphorylated tau protein and accumulated Aβ protein are considered to coexist. [17]
3D structures of amyloid precursor protein
Additional Resources
For additional information, see: Alzheimer's Disease
References
- ↑ Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008 Nov;9(11):1134-40. Epub 2008 Oct 3. PMID:18833287 doi:http://dx.doi.org/10.1038/embor.2008.188
- ↑ Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008 Nov;9(11):1134-40. Epub 2008 Oct 3. PMID:18833287 doi:http://dx.doi.org/10.1038/embor.2008.188
- ↑ Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem. 2001 Oct 26;276(43):40353-61. Epub 2001 Aug 21. PMID:11517218 doi:10.1074/jbc.M104059200
- ↑ Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001 Jun 25;153(7):1403-14. PMID:11425871
- ↑ Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The amyloid precursor protein and its regulatory protein, FE65, in growth cones and synapses in vitro and in vivo. J Neurosci. 2003 Jul 2;23(13):5407-15. PMID:12843239
- ↑ Walter MF, Mason PE, Mason RP. Alzheimer's disease amyloid beta peptide 25-35 inhibits lipid peroxidation as a result of its membrane interactions. Biochem Biophys Res Commun. 1997 Apr 28;233(3):760-4. PMID:9168929 doi:10.1006/bbrc.1997.6547
- ↑ Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001 Oct 26;276(43):40288-92. Epub 2001 Sep 5. PMID:11544248 doi:10.1074/jbc.C100447200
- ↑ Rank KB, Pauley AM, Bhattacharya K, Wang Z, Evans DB, Fleck TJ, Johnston JA, Sharma SK. Direct interaction of soluble human recombinant tau protein with Abeta 1-42 results in tau aggregation and hyperphosphorylation by tau protein kinase II. FEBS Lett. 2002 Mar 13;514(2-3):263-8. PMID:11943163
- ↑ Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009 Feb 19;457(7232):981-9. PMID:19225519 doi:10.1038/nature07767
- ↑ NCBI Reference Sequence: NP_001155.1
- ↑ UniProtKB/Swiss-Prot O00213 (APBB1_HUMAN)
- ↑ UniProtKB/Swiss-Prot O00213 (APBB1_HUMAN)
- ↑ Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001 Oct 26;276(43):40288-92. Epub 2001 Sep 5. PMID:11544248 doi:10.1074/jbc.C100447200
- ↑ Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007 Jul;3(3):186-91. PMID:19595937 doi:10.1016/j.jalz.2007.04.381
- ↑ Vivian Hook, Israel Schechter, Hans-Ulrich Demuth, Gregory Hook. Alternative Pathways for Production of Beta-Amyloid Peptides of Alzheimer's Disease.Biol Chem. 2008 August; 389(8): 993-1006. PMCID: PMC2654319
- ↑ Bharadwaj PR, Dubey AK, Masters CL, Martins RN, Macreadie IG. Abeta aggregation and possible implications in Alzheimer's disease pathogenesis. J Cell Mol Med. 2009 Mar;13(3):412-21. PMID:19374683 doi:10.1111/j.1582-4934.2009.00609.x
- ↑ Huang HC, Jiang ZF. Accumulated amyloid-beta peptide and hyperphosphorylated tau protein: relationship and links in Alzheimer's disease. J Alzheimers Dis. 2009 Jan;16(1):15-27. PMID:19158417 doi:10.3233/JAD-2009-0960
Further reading
Alzforum: Alzheimer Research Forum