Introduction
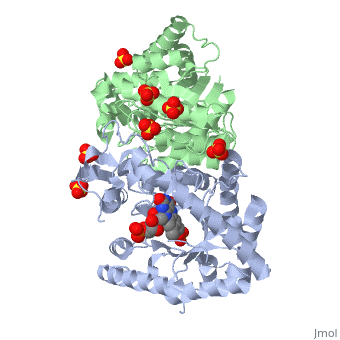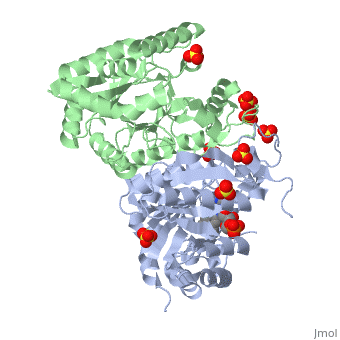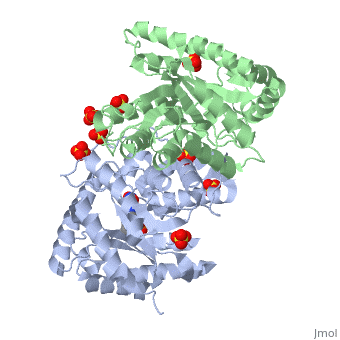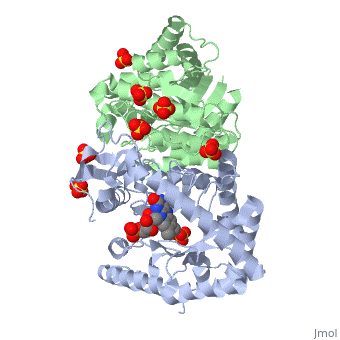
Luciferases are a class of enzymes that catalyze the oxidation of a long chain aliphatic aldehydes and emit photons. This is one type of enzyme responsible for bacterial bioluminescence. The luciferase found in Vibrio harveyi is a heterodimer that is composed of a catalytic α subunit and a homologous but noncatalytic β subunit. This reaction results in the formation of a carboxylic acid, reduced flavinmononucleotide and the emission of photons in the form of blue-green light. The catalytic α subunit houses the active site and is connected to the β subunit via a single interatcion between the mobile loop and the α subunit at α Phe 272 and Tyr 151 of the β subunit.
Mechanism of Bioluminescence
Luciferase found in V. Harveyi binds noncovalently to a reduced flavin mononucleotide cofactor, an aliphatic aldehyde and oxygen to yield oxidized flavin mononucleotide, water, and carboxylic acid. The reaction occurs in two steps forming a hydroxyflavin intermediate and ultimately results in the oxidation of the aldehyde and emission of photons[1].
FMNH2+O2+RCHO→FMN+RCOOH+H2O+hv(490nm)
The catalytic α subunit houses the FMN cofactor and is connected to the β subunit via a hairpin structure called the . The organic substrate for bacterial luciferase in vivo is myristic aldehyde, although many aliphatic aldehydes of various lengths can induce bioluminescence in vitro[2]. Oxygen is needed for light generation, no bioluminescent activity occurs in anaerobic conditions[2].
Structural Motifs
Structure homology-There is a great deal of sequence homology and structural coservation between the α and β subunits. When superimposed over the barrels of the alpha and beta subunits with a deviation of 0.62Å for 42 equivalent α carbons. The region of the beta subunit that contains the 29 residue deletion with respect to the alpha subunit differs notably in arrangement[3] . In the alpha subunit, the α7a helix is straight and extends toward the beta subunit. The region involved with dimerization, helices α and β are exceptionally similar in superposition.
Active Site and Alpha Subunit-the of bacterial luciferase is a large open cavity that is accessible to solvent via an opening located at the C-terminal ends of the ǰ strands of the TIM-barrel structure[4]. During the first step of the oxidation reaction, FMNH2 binds to the flavin binding pocket and the enzyme undergoes a conformational change. This blocks water from the surrounding environment from attacking the excited peroxydihydroflavin intermediate. Next, O2 and a long chain aldehyde bind to the FMNH2 luciferase complex and a two step oxidation reaction occurs[5].
.
The β subunit: The beta subunit is characterized as a necessary but non-catalytic subunit that stabilizes the catalytic α subunit that is responsible for the oxidation reaction. The beta and alpha subunits are connected by a single interaction between the
Mobile Loop: Residues 272-288 on the α are known as the mobile loop. This portion of the alpha subunit contains a single residue that forms a salt bridge with the beta subunit and stabilizes the active site[6].
(β/α)8 TIM Barrel- The tertiary structure of the α and β subunits are very similar, except the alpha subunit contains an extra 29 residues that the beta lacks. These 29 subunits make up the mobile loop. Both subunits fold into a single-domain eight-stranded β/α barrel motif. the two subunits assemble around a parallel four-helix bundle centered on a pseudo 2-fold axis that relates the alpha and beta subunits[7].
.