P53R2
From Proteopedia
| |||||||
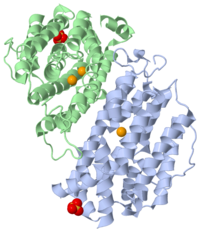
| p53R2 complex with Fe and sulfate ions, 3hf1 | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | RRM2B, P53R2 (Homo sapiens) | ||||||
| Activity: | Ribonucleoside-diphosphate reductase, with EC number 1.17.4.1 | ||||||
| Related: | 1xsm, 2uw2, 1w69, 1w68 | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
p53R2 is an oxydoreductase composed of 351 residues, that could be a new target for an anticancer therapy. It is a small subunit of the ribonucleotide reductase (RNR). RNR catalyses the reduction of the four nucleotides to desoxyribonucleotides. It exists three classes of RNR. Class I RNR is a tetramer composed of the two types of subunits with stoichiometry α2β2 and three subunits have been identified in mammals :
- Large (α) subunit M1 that contains the enzyme active site
- Small (β) subunit M2 that contains a dinuclear iron site, it’s the regulatory subunit
- P53R2, the last identified (in 2000), which is transactivated by p53 in response to DNA damage in cells during the G0-G1 cell cycle phase
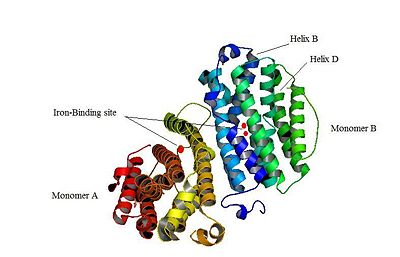
M2 and p53R2 interact with M1 through the C-terminal binding domain. These two subunits share more than 80% sequence identity. But the few differences between the two are not unimportant, as it’s explained below. The first X-ray crystal structure of p53R2 has a resolution of 2,6 Å and permits to describe its structure and also to show the structural differences with the M2 subunit (Figure 1).
Contents |
Structure and function
The X-ray crystal structure permits to see that p53R2 is made of two monomers A and B themselves made of loops and helix. Two of them play an important role. is highlighted. But concerning this site, the two monomers are not the same. Actually, the B monomer has two iron-binding site (called Fe2 and Fe1) whereas the A monomer has only one which is Fe2. This can be explained by structural changes in the helix that compose the two monomers. The 37 to 42 N-terminal residues (called the swivel loop) from one monomer can rotate between two conformations and can influence the position of the helix B or D on the opposite monomer.
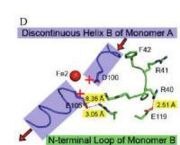
The N-terminal residues of the monomer A can stabilize the B helix of the monomer B due to different interactions. R41 of the monomer A forms a salt bridge with E119 of monomer A. This interaction permits the formation of a H-bond between R40 of monomer A with G101of monomer B. Furthermore K37 in monomer A forms a salt bridge with E105 of monomer B and this stabilize its B helix. All the interactions allow D100 of monomer B to be well oriented to bind Fe1 (see Figure 2, Smith P. et al., [1]).
On the contrary R40 of monomer B is bound to E119 of the same monomer and so it can not bind to G101 of the monomer A and the consequence is that F42 disturb the B helix of monomer A. D100 can not interact with Fe1. This explain why the monomer A has only one iron-binding site whereas the monomer B has two (see Figure 3, Smith P. et al. [2]).
Compare to the M2 subunit, these iron-binding sites are less efficient. This is due to the different conformations that the p53R2 subunit can adopt (stabilization or not of the two helix B and D). Furthermore the iron is the cofactor of the reaction catalyses by the RNR. The fact that in the p53R2 subunit the iron-binding is less efficient permits to imagine a specific anti-cancer therapy that targets these region, for example the drug deferoxamine mesylate an iron chelator. Without iron, the reduction of the nucleotides can not take place and this could avoid the proliferation of cancer cells.
Target as an anticancer therapy
The p53R2 subunit of the RR acts against cancer in three different ways: the blockage of a MAPK pathway, the repair of the DNA, and the antioxidant activity of p53R2.
- The blockage of a MAPK pathway
It has been shown that many human tumors require a MAPK pathway to progress and create metastasis. The MAPK pathway using MEK-ERK signaling is the most involved in tumor progression. Indeed, ERK 1 and 2 increase the invasiveness by activating many cytosolic and nuclear substrates which can either increase the transcription of matrix metalloprotease or decrease the transcription of tight junction proteins or disrupt the focal adhesions. The p53R2 subunit of the RR can block this pathway to decrease tumor cell invasion and metastasis.
Experiences from C Piao et al.[3] show that p53R2 and MEK2 are colocalized in the cytoplasm and that they can bind. This binding is constitutive and depends on the amino acid segment 161 to 206 of p53R2. MEK1 and MEK2 are threonine/tyrosine kinases, they can activate ERK1 and 2 by phosphorylation. The p53R2 regulates negatively the MEK-ERK pathway: it suppresses ERK activation by MEK. p53R2 inhibits tumor cells invasion and metastasis mediated by the blockage of the MEK-ERK pathway and the transcription activity.
- The DNA repair
The large subunit of the RR: R1 can either interact with R2 or with p53R2 to become catalytically active and produce desoxyribonucleotides for the DNA synthesis. p53R2 is activating by the transcription factor p53. When the cell is stressed, p53 accumulates in the cell and is subjected to post transcriptional modifications which activate the transcription of some genes. This leads to the recruitment of some proteins which will participate to DNA repair. p53R2 helps to repair the DNA by supplying desoxyribonucleotides in a cell where DNA were damaged so it prevents from mutation and cancer.
- p53R2 antioxidant activity
p53R2 is included in the RR class I which are characterized by a tyrosyl radical coupled with diiron cluster. A protein which possess tyrosyl radicals with binuclear center is known to have an oxidative or a reductive effect. So for the researchers Lijun Xue et al. [4] it raises the question of the redox properties of p53R2. They found that p53R2 is an antioxidant which can scavenge H2O2. The mitochondrial membrane is sensible to oxidative damage. H2O2 is a reactive oxygen specie (ROS) which can act on cellular growth and survival and is involved in pathogenesis like cancer. p53R2 prevents the loss of mitochondrial membrane by scavenging H2O2. This process protects cells by maintaining their genomic integrity and prevent from cancer.
3D structures of ribonucleotide reductase
References
- ↑ Smith P, Zhou B, Ho N, Yuan YC, Su L, Tsai SC, Yen Y. 2.6 X-ray Crystal Structure of Human p53R2. Biochemistry. 2009 Sep 3. PMID:19728742 doi:10.1021/bi9001425
- ↑ Smith P, Zhou B, Ho N, Yuan YC, Su L, Tsai SC, Yen Y. 2.6 X-ray Crystal Structure of Human p53R2. Biochemistry. 2009 Sep 3. PMID:19728742 doi:10.1021/bi9001425
- ↑ Piao C, Jin M, Kim HB, Lee SM, Amatya PN, Hyun JW, Chang IY, You HJ. Ribonucleotide reductase small subunit p53R2 suppresses MEK-ERK activity by binding to ERK kinase 2. Oncogene. 2009 May 28;28(21):2173-84. Epub 2009 Apr 27. PMID:19398949 doi:10.1038/onc.2009.84
- ↑ Xue L, Zhou B, Liu X, Wang T, Shih J, Qi C, Heung Y, Yen Y. Structurally dependent redox property of ribonucleotide reductase subunit p53R2. Cancer Res. 2006 Feb 15;66(4):1900-5. PMID:16488986 doi:66/4/1900
Proteopedia Page Contributors and Editors (what is this?)
Stéphanie Kraemer, Stéphanie Kilens, David Canner, Michal Harel