Sandbox Reserved 712
From Proteopedia
| |||||||||
| 3ggu, resolution 1.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | Gag-Pol, pol (Viruses) | ||||||||
| Activity: | HIV-1 retropepsin, with EC number 3.4.23.16 | ||||||||
| Related: | 3ggt | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Description
is a drug resistant HIV-1 protease. Shown is a patient's variant in complex with darunavir.
HIV proteases (PR) are essential for the functioning of the retrovirus that causes AIDS. HIV needs active proteases to process gag & gap - polymerase polyprotein precursors into mature structural proteins and replicative enzymes. HIV-1 proteases contain a highly conserved region Asp - Thr - Gly (, and ), with the aspartic residue being the in the aspartyl protease.[1]
Because of its importance for the life-cycle of the retrovirus, HIV-1 proteases are the major target for anti-HIV treatment. HIV protease inhibitors are the most potent agents used in anti-HIV treatment. However it appears that HIV-PRs develop a resistance to the inhibitor. [2]
3ggu (also PRDRV5) is a mutated clinically derived PR that shows phenotypical resistance to darunavir. Darunavir is a human immunodeficiency virus (HIV) protease (PR) inhibitor (PI) which has inhibiting effects on many HIV type 1 PR variants that show resistance to earlier-generation-PIs. [3]
Darunavir: HIV-Protease Inhibitor
Darunavir (also known as TMC114) is a HIV-PR Inhibitor. The wild-type HIV as well as a large panel of PI-resistant recombinant viruses derived from clinical samples show high susceptibility to darunavir. [4] One reason for the high potency of darunavir is the strong binding of the PI to the main-chains of the protease active-site amino acids. [5] A second reason might be the ability of the PI to fit closely into the substrate envelope. [6] Darunavir shows a high genetical barrier to resistance development. In fact studies indicate that the development of resistance needs more specific mutations and develops more slowly. [7] Darunavir resistance-associated mutations are V11I, V32I, , I47V, I50V, I54L, I54M, G73S, L76V, and (HIV Protease Resistance). Those mutations occurred in patients who have a high number of PI resistance-associated mutations. [8]
Structure
General structure of HIV-1 proteases
HIV-1 proteases are essential for HIV to survive. They are small enzymes, which are made out of two identical, 99 amino acids long, protein chains. These chains form a tunnel, which is covered by so called (flexible) "flaps". The flaps's function is to "wrap around" the substrate-protein and holding it close to the tunnel and therefore to the active site. The is located in the tunnel. The uses water molecules to break the subrate-protein chain. Its most important residues are two aspartate amino acids. Inhibitors occupy similar positions as the natural substrate-protein chains. [9]
X-ray structure analysis of 3ggu
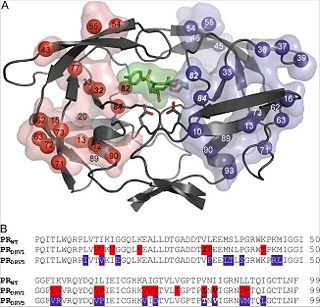
The crystal structure was determined in complex with darunavir with 1.8-Å resolutions. The crystal is formed out of one PR dimer in the asymmetric unit with two inhibitor molecules bound in alternative orientations. Surface residues and have disordered side chains, but the other amino acid residue changes could be modeled into well-defined electron density maps.
PRDRV5 contains darunavir mutations and (see Fig.1, Part B, indicated in bold print) that are directly involved in substrate-darunavir-interactions (change of S2/S2' subsites).
The other 18 mutations are outside the binding cleft, but some are still in direct contact with the binding residues (e.g. , K20M, , and ).
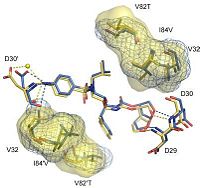
, , , , , M46I, and cause structural changes in the flap region and the flap hinge. The pictures on the right (Fig.2 and Fig.3) show the regions (indicated in blue) that undergo structural changes caused by the mutations. To see the full images, with changes in PRDRV1 and comparative structure of wild-type, PRDRV1 and PRDRV5 follow the links: Structural changes in PRDRV mutants relative to wild-type PR and Detailed view of the darunavir-enzyme interactions
It was discovered that the inhibitor substituents can adjust their positions depending on changes of the substrate binding pockets. Among them the P2' aminophenyl moiety undergoes the biggest changes. [3]
Phenotypic susceptibility and enzymatic analysis
Samples PRDRV1 to PRDRV6 have been cloned and expressed in E. coli, purified and characterized in vitro by monitoring cleavage of a chromogenic peptide substrate in the presence and absence of specific PIs.
PRDRV5 () shows twenty mutated amino acids. The PR mutations that are associated to the darunavir resistance (HIV Protease Resistance) are , and .
These mutations lead to a change in the susceptibility to the PI. In the case of 3ggu we observe a 32-fold susceptibility to darunavir. In comparison to amprenavir, which is a structural related PI of darunavir, it only shows a 24-fold susceptibility. The key-mutations that are responsible for the darunavir resistance are V32I, I54L and I54M. Those were not found in PRDRV5 which explains the smaller phenotypic changes in the susceptibility to darunavir. (Complete Table: Genotypes and phenotype changes analyzed with recombinant virus assay) Nevertheless, determining the inhibition constants by kinetic analysis using a chromogenic peptide substrate and the appropriate inhibitor, we can observe an increase of the Ki value for all the samples in comparison to the wild-type virus. PRDRV5 - which only has a specific activity of 5% of the wild-type value - also shows a smaller difference in ki value for darunavir in comparison to the other used samples. (Complete Table: Ki values for the inhibitors of PR mutants) [3]
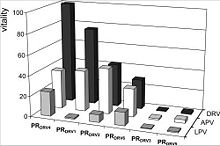
The relative vitality values are defined as v = (Kikcat/Km)MUT/(Kikcat/Km)WT. It describes the relative ability of a PR species to hydrolyze its substrate when the inhibitor is present. This means the higher the vitality the more does the mutated PR support the viral replication. [10]
The relative vitality is related to the phenotypic changes in the susceptibility to darunavir. As one can see in the diagram, the more darunavir-associated mutations there are, the higher is the relative vitality (PRDRV4 > PRDRV1 > PRDRV2 > PRDRV6). Due to the fact that PRDRV5 does not have the key mutations, it has a low vitality value for darunavir and the structural related amprenavir in comparison to the other samples. The lopinavir pattern looks different than the overall pattern of darunavir and amprenavir, because it has a different structure and resistance profile than the others.(Fig.4) [3]
Despite the many mutations the kcat values were still between 30% and 50% of the wild-type value. In contrast the Km values of the mutants were (mostly) four- to eightfold higher than the wild-type PR. (Complete Table: Enzyme characteristics of PR variants analyzed in this study) [3]
Applications
HIV (human immunodeficiency virus) which causes AIDS (Acquired immunodeficiency syndrome) is one of the most threating viruses today. The high mutation rate of the virus leads to the fast development of drug resistance. The phenotypic characterization, enzyme kinetics and X-ray structural analysis of recombinant viruses offer a way to get a better understanding of the drug resistance.[3] The knowledge we gain through that kind of experiments could help to develop new drugs for HIV-positive patients in the future.
External Resources
Human immunodeficiency virus type 1 (BRU ISOLATE)
An interactiv image that explains all the compontents of a virus
References
- ↑ Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RA, Scolnick EM, Sigal IS. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4686-90. PMID:3290901
- ↑ Watkins T, Resch W, Irlbeck D, Swanstrom R. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 2003 Feb;47(2):759-69. PMID:12543689
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Saskova KG, Kozisek M, Rezacova P, Brynda J, Yashina T, Kagan RM, Konvalinka J. Molecular characterization of clinical isolates of human immunodeficiency virus resistant to the protease inhibitor darunavir. J Virol. 2009 Sep;83(17):8810-8. Epub 2009 Jun 17. PMID:19535439 doi:10.1128/JVI.00451-09
- ↑ De Meyer S, Azijn H, Surleraux D, Jochmans D, Tahri A, Pauwels R, Wigerinck P, de Bethune MP. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob Agents Chemother. 2005 Jun;49(6):2314-21. PMID:15917527 doi:10.1128/AAC.49.6.2314-2321.2005
- ↑ Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother. 2003 Oct;47(10):3123-9. PMID:14506019
- ↑ King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J Virol. 2004 Nov;78(21):12012-21. PMID:15479840 doi:10.1128/JVI.78.21.12012-12021.2004
- ↑ De Meyer S, Azijn H, Surleraux D, Jochmans D, Tahri A, Pauwels R, Wigerinck P, de Bethune MP. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob Agents Chemother. 2005 Jun;49(6):2314-21. PMID:15917527 doi:10.1128/AAC.49.6.2314-2321.2005
- ↑ Clotet B, Bellos N, Molina JM, Cooper D, Goffard JC, Lazzarin A, Wohrmann A, Katlama C, Wilkin T, Haubrich R, Cohen C, Farthing C, Jayaweera D, Markowitz M, Ruane P, Spinosa-Guzman S, Lefebvre E. Efficacy and safety of darunavir-ritonavir at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomised trials. Lancet. 2007 Apr 7;369(9568):1169-78. PMID:17416261 doi:10.1016/S0140-6736(07)60497-8
- ↑ HIV-1 Protease, June 2000 Molecule of the Month by David Goodsell DOI: 10.2210/rcsb_pdb/mom_2000_6
- ↑ Gulnik SV, Suvorov LI, Liu B, Yu B, Anderson B, Mitsuya H, Erickson JW. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995 Jul 25;34(29):9282-7. PMID:7626598
Contributors
Julia Baaske, Angelika Wackerl

