User:Dan Huettner/Sandbox 3
From Proteopedia
Contents |
HIV-1 PROTEASE
INTRODUCTION
Acquired immune deficiency syndrome (AIDS) is arguably of the most prominent and deleterious pandemics of the modern era. AIDS is a condition in humans that causes progressive failure of the immune system, leaving individuals susceptible to opportunistic infections and cancers. The failure of the immune system results from viral assault on the human body’s white blood cells. Since the middle of the 20th century, the disease has spread rapidly in a near ubiquitous fashion, and now the world is on the brink of the fourth decade of the AIDS epidemic.

In the United States, the Center for Disease Control (CDC) first recognized AIDS during the summer of 1981.[1] Subsequently in 1983, the discovery of the sinister “AIDS virus” coined it as the human immunodeficiency virus (HIV). According to the 2010 UNAIDS Global Report, in 2009, there were 33.3 million people (adults and children) living with HIV, and 1.8 million AIDS related deaths that year, which leads to a deplorable grand total of roughly 30 million deaths due to AIDS since the HIV virus began ravaging the world.[2]
VIROLOGY
The HIV is classified in the Retroviridae family, and more specifically belongs to the genus Lentivirus – slow viruses characterized by a long incubation period, which typically result in long-duration illnesses.[3] Retroviruses (retro is the Latin prefix for “backward”) uniquely possess an RNA genome along with an RNA-dependent DNA polymerase, reverse transcriptase, which is an enzyme that can remarkably direct the synthesis of DNA from RNA.[4] HIV has been classified into two types: HIV-1 and HIV-2. The former, HIV-1, was the initially discovered form – it is more virulent and has global prevalence, making it responsible for the vast majority of infections. Conversely, HIV-2 is less virulent and mainly confined to Western Africa.[5]
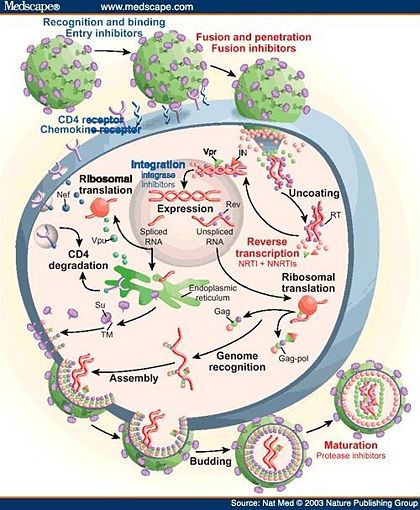
The HIV retrovirus has a fairly simple life cycle. The infection begins with the fusion of the viral and host membranes of essential immune cells, typically T-cells and macrophages. After the deposition of the viral core into the host’s cytoplasm, this capsid is uncoated, releasing viral RNA and enzymes.[6] The single-stranded RNA genome is converted into double-stranded DNA through several steps by the reverse transcriptase enzyme. The viral DNA is then transported into the host’s nucleus and inserted into the chromosome by the virally encoded integrase enzyme. Once integration has completed, the viral genome may become latent indefinitely, avoiding detection by the host’s immune system.[4] Alternatively, the viral genome can undergo transcription to new RNA and subsequent translation into proteins by the host’s machinery. Much of the viral genes are translated initially into precursor polyproteins, which are ultimately cleaved by a final key enzyme of the virus, HIV protease (HIV-1 protease is specific to the HIV-1 class). The cleavage of the polyproteins into individual proteins may occur during assembly and/or after budding of the immature virus from the host cell.[7] These final steps completed by HIV protease are absolutely critical to the maturation of the new virus.
Several fundamental characteristics allow for HIV to become such a successful and detrimental infectious agent. First, like all lentiviruses, HIV has the unique ability to replicate in non-dividing cells.[3] Second, unlike most other retroviruses, HIV kills many of the host cells it infects, rather than causing cancer, which leads to steady suppression of the host’s immune system. Third, the virus’s reverse transcriptase has an unusually high error rate for DNA replication, because the enzyme does not have proofreading exonuclease. Interestingly, the reverse transcriptase of HIV is actually around ten times more error prone than other known reverse transcriptases.[4]
Consequently, a therapeutic cure or an effective vaccine (complicated by the tendency of HIV to rapidly mutate) against HIV/AIDS has not yet been developed; therefore, the clinical management of HIV-1 infected people largely relies on antiretroviral therapy. Since the HIV retrovirus contains enzymes imperative for its life cycle that are unique from humans, these are potential inhibitory drug targets to treat infection and to prevent the progression to AIDS. The usual, most effective treatment for HIV infection is a cocktail of drugs that inhibit fusion of the virus and the key enzymes reverse transcriptase, integrase, and protease. This approach for treatment as a combination of drugs is known as highly active antiretroviral therapy (HAART), which has improved the lives of millions around the world who suffer from HIV/AIDS.[8]
STRUCTURE AND ACTIVE SITE OF HIV-1 PROTEASE
The first structures of HIV-1 protease were reported in 1989 revealing its homodimeric structure consisting of two identical monomers, each made up of 99 amino acids residues, related by a two-fold axis of symmetry.[9] The secondary structure of each monomer contains antiparallel beta-sheets and a single alpha-helix. Each monomer is stabilized in a hydrophobic core by aliphatic residues – open long-chain carbons structures, while the homodimer is stabilized by non-covalent and hydrophobic interactions of the side chains.[10]
Flexible flaps form from an extended turn of a beta sheet (beta hairpin loop) that covers the active site.[10] Incidentally, the active site tunnel is normally too narrow for the polyprotein to fit. However, the solution is the two flexible protein flaps (amino acid resides 45-55 on each monomer) that can move to allow the peptide to enter the tunnel active site.[11] These flaps undergo a dramatic conformational change from open to closed states to bind the substrate in the proper conformation for catalytic cleavage. The highly flexible tips of the flaps are glycine rich, which curl inside the cleft as the tunnel expands, burying many hydrophobic residues and exposing electronegative active site, while widening the tunnel enough for the substrate to enter.[11]
ASPARTYL PROTEASE MECHANISM
HIV-1 protease is an aspartyl protease, similar to pepsin, which contains two essential aspartate (Asp25) residues, one from each monomer, that play a key role in the enzyme’s catalytic function at the active site. Like all aspartyl protease, the active site of HIV contains a highly conserved catalytic triad of three amino acid residues, Asp25-Thr26-Gly27 (Aspartate-Threonine-Glycine).[12] Each monomer contributes a single Asp-Thr-Gly triad to the active site, amounting to six amino acids in the active site of HIV-1 protease. The active site triads are located in a loop whose structures are stabilized by a network of hydrogen bonds. It has been hypothesized that the role of the Thr26 residues is to stabilize the conformational state of the active site through hydrogen bonding forces with one another, also known as “fireman’s grip”.[12] Moreover, the Gly27 residues function to accommodate and bind the substrate for subsequent attack by the Asp residues.[13]
The Asp25 residues are nearly coplanar and interactive directly with the substrate. Each Asp25, being negatively charged amino acids, holds a water molecule, crucial for catalysis, through hydrogen bonding.[13] The Asp25 residues and water together cleave the substrate through a general acid/base hydrolysis reaction. The active site aspartyl residues assume opposite roles during catalysis.[12] One Asp25 residue behaves like an acid, donating a proton to the carbonyl oxygen of the substrate, as the other Asp25 behaves like a base, accepting a proton from water. This promotes the nucleophilic attack by the water on the substrate, cleaving the peptide bond of the polyprotein.[14]
DRUG INHIBITORS AS TREATMENTS
The active site of HIV-1 protease has been one of the key targets for fighting HIV/AIDS. The drug inhibitors are highly stable mimickers of the polyproteins that cannot be cleaved by HIV-1 protease. They are able to bind with higher affinity than the polyprotein substrate. These drugs function as reversible inhibitors that compete with the usual substrate for the enzyme’s active site through competitive inhibition.[10]
Presently, nine protease inhibitors have been approved for clinical treatment of HIV infection: saquinavir, indinavir, ritonavir, nelfinavir, amprenavir, lopinavir, atazanavir, darunavir, and tipranavir.[12] These inhibitors are smaller in size to the intended peptide substrates. They contain a central hydroxyl group that mimics the tetrahedral reaction intermediate, interacting with the carboxyl groups of the Asp25 residues of the active site, increasing their affinity for protease. Additionally, all the inhibitors contain polar groups that create hydrogen bonding interactions with numerous other residues within the active site tunnel, which are mediated by a conserved water molecule.[10] Figure 6 shows an inhibitor, Indinavir (PDB 1HSG), tightly bound to the active site of HIV-1 protease.
Protease inhibitors have been fundamental components of treatment therapies for HIV/AIDS since 1995, when antiviral protease inhibitors were first approved for clinical use. The integration of protease inhibitors in drug therapy has been associated with successful therapeutic treatment of HIV/AIDS, significantly reducing AIDS mortality and morbidity.
=REFERENCES
- ↑ Gallo, Robert C. (2002). The early years of HIV/AIDS. Science, 298(5599), 1728-1730.
- ↑ UNAIDS, World Health Organization (WHO). (2010). Global Report: UNAIDS Report on the Global AIDS Epidemic: 2010. Retrieved April 26, 2011.
- ↑ 3.0 3.1 Lévy, J. A. (1993). HIV pathogenesis and long-term survival. AIDS, 7(11), 1401–10.
- ↑ 4.0 4.1 4.2 Nelson, David L., & Cox, Michael M. Lehninger Principles of Biochemistry, Fifth Edition; W. H. Freeman and Company: New York, 2008; pp 218-219, 1050-1053.
- ↑ Gilbert, PB et al; McKeague, IW; Eisen, G; Mullins, C; Guéye-Ndiaye, A; Mboup, S; Kanki, PJ (2003). Comparison of HIV-1 and HIV-2 infectivity from a prospective cohort study in Senegal. Statistics in Medicine, 22(4), 573–593.
- ↑ Campbell, E.M., & Hope, T.J. (2008). Live cell imaging of the HIV-1 life cycle. Trends Microbio., 16(12), 580-587.
- ↑ Gelderblom, H. R (1997). Fine structure of HIV and SIV. HIV Sequence Compendium. Los Alamos, New Mexico: Los Alamos National Laboratory. pp. 31–44.
- ↑ Lu, X. F., & Chen, Z. W. (2010). The development of anti-HIV-1 drugs. Yao Xue Xue Bao = Acta Pharmaceutica Sinica, 45(2), 165-176.
- ↑ Spinelli, S.; Liu, Q.Z.; Alzari, P.M.; Hirel, .PH.; & Poljak, R.J. (1991). The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochimie., 73(11), 1391-1396.
- ↑ 10.0 10.1 10.2 10.3 Louis, J. M., Ishima, R., Torchia, D. A., & Weber, I. T. (2007). HIV-1 protease: Structure, dynamics, and inhibition. Advances in Pharmacology (San Diego, Calif.), 55, 261-298.
- ↑ 11.0 11.1 Toth, G., & Borics, A. (2006). Flap opening mechanism of HIV-1 protease. Journal of Molecular Graphics & Modelling, 24(6), 465-474.
- ↑ 12.0 12.1 12.2 12.3 Castro, H. C., Abreu, P. A., Geraldo, R. B., Martins, R. C., dos Santos, R., Loureiro, N. I., Cabral, L.M., &Rodrigues, C. R. (2011). Looking at the proteases from a simple perspective. Journal of Molecular Recognition : JMR, 24(2), 165-181.
- ↑ 13.0 13.1 Mager, P. P. (2001). The active site of HIV-1 protease. Medicinal Research Reviews, 21(4), 348-353.
- ↑ Prashar, V., Bihani, S., Das, A., Ferrer, J. L., & Hosur, M. (2009). Catalytic water co-existing with a product peptide in the active site of HIV-1 protease revealed by X-ray structure analysis. PloS One, 4(11), 7860.