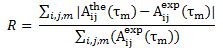User:R. Jeremy Johnson/NMR
From Proteopedia
Contents |
Introduction
Overview
Ribonuclease A has served as a model for protein structure and finction, and was the third enzyme whose three-dimensional structure was determined.[1] Ribonuclease has been called the most studies enzyme of the 20th century due to its ample availability as well as its significant role within the cell.[1] While the structures of bovine pancreatic ribonuclease (RNase A) and human pancreatic ribonuclease (RNase 1) determined by X-Ray crystallography have been around for some time, the 3D NMR structures of present provide much more information on specific locations of side chain residues and their flexibility. Because NMR does not require a "frozen" crystal structure, NMR imaging can show much more accurate detail into the actual, solution enzyme (folding, flexibility etc.)
Ribonuclease A and Ribonuclease 1 are both good targets for 3D NMR. Not only are they small proteins (~13 KDa), they also have numerous characteristics that are observable only by NMR, such as internal flexibility and 3D domain swapping.
3D NMR spectroscopy has shed light on protein folding dynamics as a whole, suggesting a framework model of folding. In other words, Ribonuclease NMR studies have provided evidence that secondary structure within the protein fold before the tertiary folds begin (Folding order = primary structure, secondary structure, tertiary structure).
NMR Versus X-Ray Crystallography
The two predominant methods of protein tertiary structure determination are X-Ray Crystallographyand Nuclear Magnetic Resonance(NMR) Spectroscopy.

To complete an X-Ray Crystallographic structure of a protein requires protein purification, crystallization of the protein, collection of X-Ray diffraction data, calculation of the protein’s electron density, and finally fitting the protein’s determined amino acid sequence into the electron density. Crystallizing the protein is often the rate-limiting step in structure determination. While any size protein can be studied via X-Ray Crystallography and the method is well-established, it is often difficult to crystallize membrane proteins, hydrogen atoms are not present, and crystal conditions are assumed to mimic solution conditions. For more information regarding X-Ray Crystallography please click here.
To complete a bi-molecular NMR Spectroscopy structure of a protein requires protein purification, dissolving the protein in a suitable solvent, collecting the NMR data, assigning NMR signals, and finally calculating the protein’s tertiary structure. With NMR, the most difficult step is often correctly assigning NMR signals. Although there is no need to crystallize the protein of interest and most hydrogen atoms are present, NMR is difficult for proteins that do not dissolve well in common solvents and works best for small proteins. 1-dimensional, 2-dimensional, and 3-dimensional NMR spectroscopy is readily available; however, 2-dimensional and 3-dimensional NMR are most often used for protein tertiary structure determination. 2D NMR reveals chemical shift correlations between spinnable nuclei such as 1H, 13C, 15N, and 13P, as well as atomic coupling, or proximity correlations via bonding. 3D NMR utilizes this methodology in addition to detection of another nuclear spin phenomenon known as the Nuclear Overhauser Effect(NOE), in which proximity correlations are can be observed in 3D space.

Due to the complexity of assigning NMR signals to specific protons, complex computer algorithms are required to render a protein’s tertiary structure. Even though NMR often requires more prior knowledge of the studied protein’s structural information, the end result contains more information about the solution structure of the protein including the protein’s conformational flexibility (see . For more information regarding NMR Spectroscopy, click here.
History of NMR Ribonuclease Studies
In 1957, Martin Saunder et al examined the structure of bovine pancreatic Ribonuclease using 1-Dimensional 1H NMR.[4]
In 1988, Udgaonkar et al. used 2-dimensional 1H NMR to study bovine pancreatic Ribonuclease and measured protein folding dynamics, which supported the framework model protein folding mechanism.[5]
In 1993, Santoro et al. used 3-dimensional 1H NMR to study bovine pancreatic Ribonuclease to compare NMR structures with X-Ray Crystallography structures of RNase A.[6]
In 2008, Rico et al. used 3-dimensional 1H NMR to study human pancreatic Ribonuclease. They looked at active site conformational changes upon substrate binding, and suggested possible biological implications.[7]
Medical Significance
Ribonucleases show specific toxicity to tumor cells. In fact, Onconase ® (an RNase A homolog from the Northern Leopard Frog) is currently in phase IIIb clinical trials for the treatment of malignant mesothelioma. However, due to possible immunogenicity of the frog enzyme, much effort has been focused on developing a cytotoxic human RNase, which evades inhibition by human ribonuclease inhibitor (HRI). HRI selectively binds to human pancreatic RNase (RNase 1) and impedes its enzymatic activity. Correctly characterizing the structure and binding specificity of RNase via NMR could assist in the development of RNase-based anti-cancer treatments.
NMR Study of Ribonuclease Folding Dynamics[5]
Experimental Procedure
Using 2-dimensional 1H NMR, Udgaonkar et al. studied the folding pathway of bovine pancreatic Ribonuclease using an exchange reaction between with solvent protons. 2- dimensional 1H NMR allowed for monitoring of proton exchange in the amide backbone for ten second time intervals, and this proton labeling could be terminated via a rapid drop in pH reaction conditions. This research focused on the initial protein folding steps.
Starting with denatured wild-type RNaseA, it was hypothesized that as the peptide began to fold, the backbone amide proteins would become less energetically favorable to exchange protons with the solvent as the backbone amide protons became involved in folding-related intermolecular interactions (such as ).
Data and Results
were shown to be involved in folding-related intermolecular interactions during initial protein folding steps: amide protons from , , , , and . All five of these protons are involved in hydrogen bonding within the of Ribonuclease; therefore, β sheets were proposed to be the starting point for the folding mechanism of Ribonuclease. Furthermore, this supports the formation of a stable secondary structure before the formation of the , which is consistent with the framework model of protein folding mechanisms.
|
Ribonuclease NMR Structure Versus X-Ray Crystallography Ribonuclease Structure[6]
Experimental Procedure
Using 3-dimensional 1H NMR (2D NMR with NOE enhancement), Santoro et al. studied the tertiary structure of bovine pancreatic Ribonuclease. Their experimental raw data were processed by TRITON software, and previously determined proton assignments were utilized. The NMR tertiary structure results were categorized by their quality, or by the size and number of residual distance constraint violations. Using an equation from X-Ray Crystallography studies, the reliability factor (R-Factor) of each structure can be defined by comparing experimental NOE intensities with theoretical NOE intensities. The R-Factor can be defined as:
Where A(the) is the theoretical NOE intensity and A(exp) is the experimentally determined NOE value.
Data and Results
The NMR experiment yielded an overall R-Factor of 0.44, compared to an X-Ray Crystallographic R-Factor of 0.45. This means that the NMR structure shows a higher structural reliability compared to the X-Ray Crystallographic structure. Overall, the NMR tertiary structure of RNase matches closely with the corresponding X-Ray crystallography structure. The overall shape, main-chain fold, and side chain positions of most residues are similar between the two structures. Experimentally determined tertiary structural differences between the two methods were suggested to be due to pH differences, crystal packing, solvation, and temperature variability.
Previously, researchers found the side chain position of in the enzyme’s () of NMR structures to be different than that of X-Ray Crystallography studies. Crystals show a static position of this His 119 residue, yet NMR structures suggest a dynamic equilibrium between the two conformational puckers of the . This single residue difference between crystal and solution studies amplifies to cause a major difference in surrounding amino acid residues: , , and . The researchers proposed that this difference is most likely due to pH induced charge repulsion of His 119 with and in solution.
More than 60 main-chain hydrogen bonds were observed, which closely corresponds to the number of hydrogen bonds determined in crystals; however, a few discrepancies existed such as hydrogen bonds between the amide proton on , , , , and . The researchers suggested these differences are most likely due to the same pH phenomenon mentioned above.
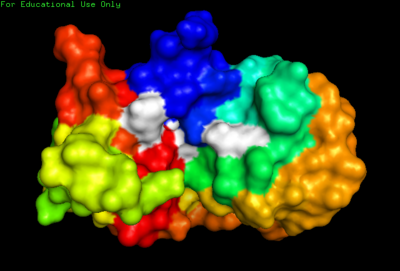
The NMR structure also highlighted flexibility of RNase A. Overall, the largest conformational flexibility was found in the side-chains. Specifically, side-chain mobility is greatest in residues (shown in white). As expected, the backbone torsion angles were seen to be more rigid (less conformational flexibility) within the of RNase A.
Solution Structure and Dynamics of Human Pancreatic Ribonuclease[7]
|
Experimental Procedure
Using 3D NMR, Rico et al. obtained the first NMR 3-dimensional structures of Human RNase 1. Using 20 different RNase 1 NMR structures, researchers compared the RMSD values of key RNase 1 residues. A residue with a high RMSD value has more flexibility and a residue’s ability to adapt to numerous conformations may be crucial to its active role within the enzyme.
Furthermore, this study characterized the dimerization of wt Ribonuclease and mutated Ribonuclease variants. Similar dimerization, observed in certain Bovine RNases, greatly enhances enzymatic activity and anti-tumoral action. Using varying enzyme concentrations, pH conditions and site-directed mutations, RNase 1 was dimerized.
This study also compares the location of certain residues in bound RNase 1 to unbound RNase. This comparison, provides greater insight into the catalytic mechanism and substrate specificity of RNase 1.
Data and Results
A 3D NMR structure was obtained with a backbone RMSD of 1.07Å. The obtained model shows a similar tertiary structure to the kidney bean shaped RNase A and is stabilized by four . The structure shows three and seven . While this structure matches up fairly well with previous X-Ray crystallography structures of RNase 1, important differences in residue positioning can be seen in the which are not apparent in X-Ray crystallography. Specifically, certain residues with more flexibility undergo a significant conformational change when bound to certain substrates, such as the human ribonuclease inhibitor (hRI). These residues include: .
This large conformational change suggests an “induced-fit” model of substrate binding and may prove vital to fully understanding RNase 1’s binding specificity for Hcrl; although two residues, , show much more rigidity and possibly contribute some “lock-and-key” binding interaction. Designing cytotoxic variants of RNase1 may prove difficult due to the large number of active amino acids; numerous mutations may be required to fully "kill" the enyzme.
15N NMR relaxation shows increased T1 values for the residues found in these sheets and loops (0.63-0.64s relative to 0.60s in helices), suggesting greater flexibility in these regions as well.
Interestingly, the global correlation time of RNase 1 was much longer than expected for a 13.7 kDa monomer (10ns compared to 6-7ns) indicating the RNase1 undergoes dimerization forming a monomer/dimer equilibrium. Because dimerization affects the enzymatic activity of bovine ribonuclease, RNase 1 variants with increased dimerization may be potential anti-cancer therapeutics.
References
- ↑ 1.0 1.1 Raines, Ronald T. "Ribonuclease A." Chemical Reviews; 98, 1045-1065 (1998). Print.
- ↑ Dahl, Jeff. X-ray diffraction pattern of crystallized 3Clpro, a SARS protease. (2.1 Angstrom resolution). 2006. Web. 1 Apr. 2011.
- ↑ English Wikipedia. COSY NMR spectrum progesterone. Web. 1 Apr. 2011.
- ↑ Saunders, Martin, Arnold Wishnia, and John G. Kirkwood. "The Nuclear Magnetic Resonance Spectrum of Ribonuclease." Communications to the Editor 79; 20 May (1957). Print.
- ↑ 5.0 5.1 Udgaonkar JB, Baldwin RL. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694-9. PMID:2845278 doi:http://dx.doi.org/10.1038/335694a0
- ↑ 6.0 6.1 Santoro J, Gonzalez C, Bruix M, Neira JL, Nieto JL, Herranz J, Rico M. High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy. J Mol Biol. 1993 Feb 5;229(3):722-34. PMID:8381876 doi:http://dx.doi.org/10.1006/jmbi.1993.1075
- ↑ 7.0 7.1 Kover KE, Bruix M, Santoro J, Batta G, Laurents DV, Rico M. The solution structure and dynamics of human pancreatic ribonuclease determined by NMR spectroscopy provide insight into its remarkable biological activities and inhibition. J Mol Biol. 2008 Jun 20;379(5):953-65. Epub 2008 Apr 25. PMID:18495155 doi:10.1016/j.jmb.2008.04.042
- ↑ Willow. Top view of ribbon diagram of ribonuclease inhibitor (PDB accession code 2BNH). Made with MOLMOL. 2006. Web. 1 Apr. 2011..