From Proteopedia
proteopedia linkproteopedia link
|
|
| 1h6v, resolution 3.00Å ()
|
| Ligands:
| ,
|
| Non-Standard Residues:
|
|
| Activity:
| Thioredoxin-disulfide reductase, with EC number 1.8.1.9
|
| Structural annotation:
|
| Resources:
| CATH : 1H6Va03, 1H6Va01, 1H6Va02, 1H6Vb03, 1H6Vb01, 1H6Vb02, 1H6Vc01, 1H6Vc02, 1H6Vc03, 1H6Vd01, 1H6Vd02, 1H6Vd03, 1H6Ve01, 1H6Ve02, 1H6Ve03, 1H6Vf01, 1H6Vf02, 1H6Vf03
InterPro : Ipr004099, Ipr012999, Ipr001100, Ipr006338, Ipr013027, Ipr001327, Ipr000815
Pfam : PF02852, PF00070, PF07992
SCOP : d1h6va1, d1h6va3, d1h6va2, d1h6vb1, d1h6vb2, d1h6vb3, d1h6vc3, d1h6vc1, d1h6vc2, d1h6vd1, d1h6vd2, d1h6vd3, d1h6ve1, d1h6ve3, d1h6ve2, d1h6vf1, d1h6vf3, d1h6vf2
UniProt : O89049
|
|
|
|
|
|
| Resources:
| FirstGlance, OCA, PDBsum, RCSB
|
| Coordinates:
| save as pdb, mmCIF, xml
|
Thioredoxin Reductase
Thioredoxin reductase (TR) is a 55 kDa enzyme that belongs to the family of pyridine nucleotide disulfide oxidoreductases
[1]. Also included in this family are lipoamide dehydrogenases and
glutathione reductases, with which TRs share high homology. TR is ubiquitous from humans to archaea, however TR from higher eukaryotes is distinct from its prokaryotic counterpart and is thought to have evolved from gluthathione reductases due to similarities in the catalytic activity of both enzymes
[2]. Compared with gluthathione reductases, TRs are unique in that they contain a 16 amino acid C-terminal extension which mediates the catalytic activity of the enzyme
[3]. Mammalian TRs fall into the class of selenium containing enzymes due to the presence of its penultimate selenocysteine residue that has been shown to be essential for reduction of its cognate substrate,
thioredoxin. Selenocysteine is encoded by the UGA stop codon, which has to be recoded as a sense codon as shown in the figure below. The C-terminal redox center (which contains the selenocysteine residue) is notable because a number of high molecular weight TRs contain cysteine in place of selenocysteine (Sec) at this site
[4]. TR from
Drosophila melanogaster falls under this category and has a vicinal cysteine dyad in the redox center. Mammalian TR is known for having decreased catalytic activity upon replacement of the selenocysteine residue with cysteine
[5]. This aspect together with other findings not mentioned here suggests that selenocysteine plays a special role in the enzyme. For this reason, the role of selenocysteine in TR is controversial considering that cysteine is the functional residue in other forms of the enzyme.
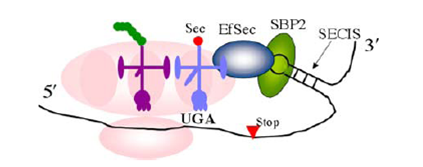
Hondal, R.J. (2010)
Amino Acids, E Pub
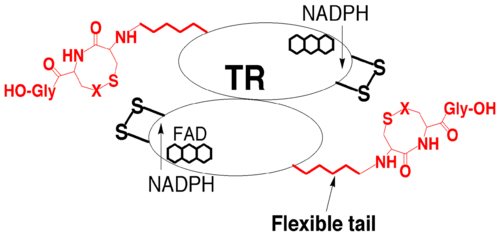
Hondal, R.J. (2010)
Amino Acids, E Pub
Structure
The functional unit of TR is a , typical of proteins in the family of glutathione reductases, with each subunit composed of mainly (yellow) and (blue). Each monomer exhibits a three domain modular architecture, containing a NADP binding domain, a N-terminal FAD binding domain, and an interface domain. Both the (in red and blue respectively) binding domains have similar folds, and are variants of the , characterized by a β sheet linked by several alpha helices which in the enzyme is composed of 5 strands surrounded by helices. The two domains are positioned in a head to tail orientation allowing for electron transfer that leads to the reduction of the enzyme’s redox active center. The of the enzyme, characterized by the conserved Cys-Val-Asn-Val-Gly-Cys sequence, is located at the interface domain formed by two subunits and reduces the C-terminus of the oppposite TR subunit, highlighting the physiological significance of the dimeric form of the enzyme. This domain is composed of a five stranded β sheet flanked on either side by two helices. The C-terminal extension of TR runs parallel to the edge of the β sheet strand at the interface domain, with the last residues of the extension forming an arm that protrudes into the interface domain allowing for interaction with groups at the active site interface, which is located at the N-terminus [6]. The isoalloxazine ring of , which is noncovalently bound to the enzyme, packs against the disulfide bond between Cys-59 and Cys-64 (numbering from the rat enzyme) within the active site with the sulfur atom of Cys64 being closest to the ring. This form of packing suggests that this residue is responsible for forming the thiolate-flavin charge transfer complex. The residues that are involved in the interaction between FAD and TR are mainly from the FAD binding domain and are highly conserved in the family of pyridine nucleotide disulfide oxidoreductases. The coenzyme NADPH binds within a cleft between the FAD and NADP binding domain with its adenine moiety packed against the central β-sheet of the NADP binding domain. The remainder of the molecule extends toward the isoalloxazine ring of the FAD.
and
are both involved in the recognition of NADPH versus NADH and form hydrogen bonds with the 2' phosphate of NADPH. The substrate binding site of TR is highly conserved within the glutathione reductase family and is comprised of four helices from one subunit and the C-terminal part of the other subunit. However, there is no experimental evidence suggesting which residues participate in the binding of thioredoxin to TR. The conservation between TR and glutathione reductase once again highly suggests that TR have evolved from glutathione reductase.
Function and Mechanism
High molecular weight TRs catalyze the reduction of the redox active disulfide of thioredoxin, the enzyme’s cognate substrate. Together with thioredoxin and NADPH, TR forms the thioredoxin system which plays a major role in maintaining a reducing environment within cells. Studies on thioredoxin have provided a vast amount of information on the function and mechanism of TR. Although the enzyme reduces disulfide containing substrates, it has a broad substrate spectrum and also targets other nondisulfide substrates such hydrogen peroxide and selenite
[7]. The general mechanism of the enzyme is initiated upon transfer of electrons from NADPH via a bound FAD to the N-terminal redox active site (CVNVGC)
[8]. One of the two cysteine residues in the N-terminal redox center forms a charge transfer complex with the FAD, while the other initiates a thiol-disulfide exchange step resulting in the reduction of the C-terminal disulfide/selenylsulfide (Gly-Cys-Sec-Gly) of one TR monomer by the N-terminal redox center of the opposite monomer. Once the C-terminus is reduced, the attacking nucleophile initiates attack on the disulfide of thioredoxin. The model below depicts the mechanism of a cysteine TR during catalysis with the thioredoxin substrate. The cysteines of the N-terminal redox center are labeled charge transfer (CT) and interchange (IC). According to Arner et al, the concept of the TR catalytic principle can be rationalized in terms of “yin and yang dualism”
[9]. This view was expressed as both subunits are aligned in a head to tail fashion and are required for electron transfer leading to thioredoxin reduction as shown in the figure below (ying yang depiction). The N-terminal dithiol-containing active site on one subunit in TR reduces the C-terminal selenosulfide/disulfide on the opposite subunit as shown below. The resulting selenothiol/dithiol motif may subsequently reduce thioredoxin and several other substrates. The Sec containing motif constitutes the active site of the enzyme and further explains the selenium dependency of TR. Mutation of this residue to a serine or truncation of the region containing the Sec residue leads to the inability of the enzyme to reduce thioredoxin, while substitution of the Sec residue with Cys leads to decreased activity towards substrate reduction
[10]. These findings raise questions concerning the role of the Sec residue in the enzyme given that it provides a catalytic advantage over Cys containing enzymes. The current notion behind Sec failing to replace Cys in enzymes is perhaps the cumbersome recoding mechanism used in translating the Sec encoding UGA codon as a sense codon for selenocysteine, which is further complicated by the fact the both eukaryotes and prokaryotes have different UGA recoding mechanisms
[11] [12]. The intricacy behind this process suggests that Sec plays an important role in the enzyme that a Cys residue is unable to fulfill, therefore a number of rationales have been proposed based on the fact that Sec is implicated in what is thought to be the rate determining step of the enzyme. Currently, the conventional wisdom is that Sec exists due to its ability to act as a superior nucleophile based on the data described earlier. At neutral pH, Sec has a lower pKa (5.2) compared to Cys (pKa-8.00), implying that Sec is significantly more nucleophilic, however, since individual rate constants for the TR mechanism have never been measured, experimental evidence supporting this notion of nucleophility is nonexistent. Another school of thought is the better leaving group potential of a selenol (SeH) relative to a thiol (SH) due to the significantly lower pKa (5.2) of Sec, which confers stability upon selenolate (Se-) in the absence of a proton which would typically be required as the selenosulfide bond is being broken. Furthermore, the acidity of selenol is much greater than that of a typical thiol due the low pka. This view suggests that the rate limiting step involves the transfer of electrons from the N-terminal redox center to the C-terminal selenosulfide site. This was determined by studying a truncated form of the enzyme missing the last 8 residues and using a synthetic peptide containing the selenosulfide in both the cyclic and linear form as a substrate for the enzyme. Based on these studies, ring geometry was found to be more important in the Cys containing enzymes versus the Sec containing mammalian enzyme. In addition, the interpretation of selenolate not requiring protonation prior to breakage of the selenosulfide bond was attributed to the fact that Sec was found to be more important in flow of electrons from the N-terminal redox center to the Sec containing motif, therefore Se is needed in the step before nucleophilic attack on thioredoxin. Recently, other views have been proposed with regard to the role of selenium in TR based on the fact that Se in a selenosulfide bond expresses a much stronger electrophilic character compared to S in a disulfide bond.
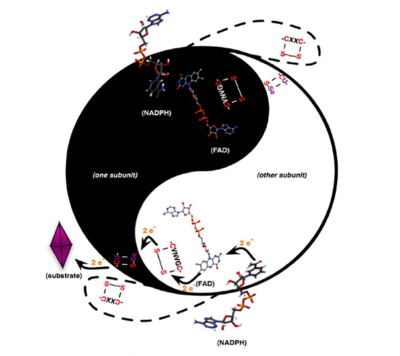
Arner, E.S.J (2009)
Biochimica et Biophysica Acta 1790,495-526
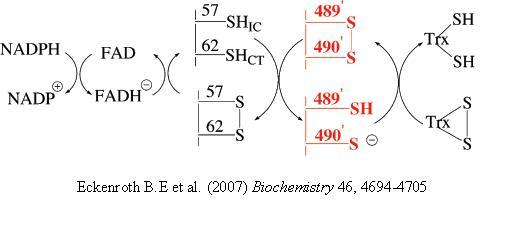
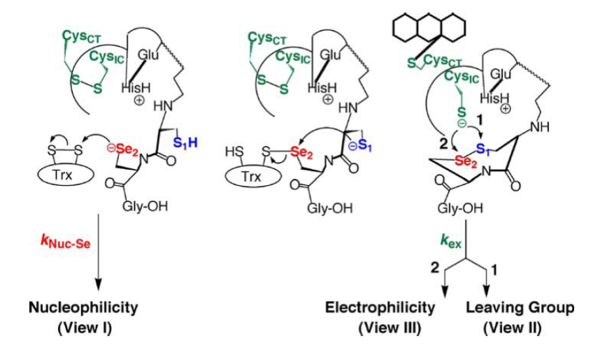
Hondal, R.J. et al (2010)
Amino Acids, E pub
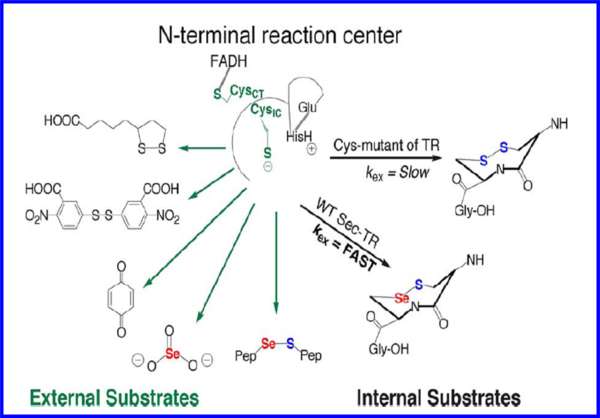
Hondal, R.J. et al (2010)
Amino Acids, E pub
Role of TR in Cardiovascular Development and Cancer
TRs are highly important for control of apoptosis, antioxidant defense, and also have multifaceted roles in cells including implications in cancer. There are three classes of TR in mammals- cytosolic TR, mitochondrial TR, and a testis-specific GSH-T. However, little is known about their individual contributions [13]. During development, mitochondrial TR is highly expressed in the heart and liver, organs involved in high metabolic activity, which further supports the crucial role of TR in the control of intracellular reactive oxidative species. Studies on mouse embryo have shown that deletion/knockout of mitochondrial TR leads to death at embryonic day 13 [14]. The embryos were found to be significantly anemic and smaller than their WT counterparts (see thumbnail) and showed increased apoptosis in the liver. Also, the ventricular heart walls were thinner and cardiomyocyte proliferation was decreased. The phenotype observed in the TR knockout mice has been observed in Keshan_disease, which is a selenium deficiency disease that affects heart function. Knowledge of this disease was the first indication that selenium is essential for heart function. In this same study, hematopoietic colonies were also found to be significantly reduced suggesting their dependence on TR. These results indicate that TR plays a significant role not only in heart function, but also in hematopoietic development.
The TR system is also involved in separate aspects within the hallmark of cancer. The involvement of TR in antioxidant defense aids in the elimination of carcinogenic oxidants and in the repair of oxidized proteins[15]. Similarly, the reduction of selenite by TR to selenide decreases oxidative stress and may contribute to certain therapeutic effects. TR is a target for existing chemotherapy drugs given its wide substrate specificity and likely involvement in ameliorating cancer during early stages. TR has also been found to be upregulated in some cancers because it provides reducing equivalents for ribonucleotide reductase thereby functioning in the synthesis of deoxyribonucleotides for DNA synthesis.
Links
- 1zkq (Mouse TR)
- 2nvk (TR from Drosophila melanogaster)
- 2cfy (Human TR)
- 1gra (Structurally related enzyme-Glutathione Reductase)
References
- ↑ Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9533-8. Epub 2001 Jul 31. PMID:11481439 doi:10.1073/pnas.171178698
- ↑ Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5854-9. PMID:10801974 doi:10.1073/pnas.100114897
- ↑ Eckenroth BE, Lacey BM, Lothrop AP, Harris KM, Hondal RJ. Investigation of the C-terminal redox center of high-Mr thioredoxin reductase by protein engineering and semisynthesis. Biochemistry. 2007 Aug 21;46(33):9472-83. Epub 2007 Jul 28. PMID:17661444 doi:10.1021/bi7004812
- ↑ Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9533-8. Epub 2001 Jul 31. PMID:11481439 doi:10.1073/pnas.171178698
- ↑ Eckenroth BE, Rould MA, Hondal RJ, Everse SJ. Structural and biochemical studies reveal differences in the catalytic mechanisms of mammalian and Drosophila melanogaster thioredoxin reductases. Biochemistry. 2007 Apr 24;46(16):4694-705. Epub 2007 Mar 27. PMID:17385893 doi:10.1021/bi602394p
- ↑ Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9533-8. Epub 2001 Jul 31. PMID:11481439 doi:10.1073/pnas.171178698
- ↑ Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9533-8. Epub 2001 Jul 31. PMID:11481439 doi:10.1073/pnas.171178698
- ↑ Eckenroth BE, Rould MA, Hondal RJ, Everse SJ. Structural and biochemical studies reveal differences in the catalytic mechanisms of mammalian and Drosophila melanogaster thioredoxin reductases. Biochemistry. 2007 Apr 24;46(16):4694-705. Epub 2007 Mar 27. PMID:17385893 doi:10.1021/bi602394p
- ↑ Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim Biophys Acta. 2009 Jun;1790(6):495-526. Epub 2009 Feb 11. PMID:19364476 doi:10.1016/j.bbagen.2009.01.014
- ↑ Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000 Jun 16;275(24):18121-8. PMID:10849437 doi:10.1074/jbc.M000690200
- ↑ Hondal RJ, Ruggles EL. Differing views of the role of selenium in thioredoxin reductase. Amino Acids. 2010 Feb 21. PMID:20397034 doi:10.1007/s00726-010-0494-6
- ↑ Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim Biophys Acta. 2009 Jun;1790(6):495-526. Epub 2009 Feb 11. PMID:19364476 doi:10.1016/j.bbagen.2009.01.014
- ↑ Sun QA, Kirnarsky L, Sherman S, Gladyshev VN. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc Natl Acad Sci U S A. 2001 Mar 27;98(7):3673-8. Epub 2001 Mar 20. PMID:11259642 doi:10.1073/pnas.051454398
- ↑ Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004 Nov;24(21):9414-23. PMID:15485910 doi:24/21/9414
- ↑ Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006 Dec;16(6):420-6. Epub 2006 Oct 28. PMID:17092741 doi:10.1016/j.semcancer.2006.10.009







