User:Wally Novak/Sandbox Miller
From Proteopedia
|
DNA ligases are a group of proteins responsible for the joining DNA fragments which arise from various cellular events: ligating Okazaki fragments from replication, various repair mechanisms, and recombination.[1] The ligation proceeds via a three step process uniting the 3'-OH to the 5'-phosphate with an ATP or NAD⁺ cofactor.[2] DNA ligase I is a member of this family expressed throughout eukaryotes. Included is a summary of its functionality covering the conserved structural domains, ligating mechanism, and involvement in replication as well as DNA repair mechanisms.
Contents |
Structural Domains
DNA ligase I has four structural domains spread over a 919 residue monomeric protein. Three of the four domains are highlighted in the . The DNA-binding domain (DBD) is in red; the adenylation domain (AdD), green; the OB-fold domain (OBD), yellow; and the DNA, grey. The first 232 residues containing domains that interact with other regulative proteins were excluded from the source structure that Ellenberger and colleagues studied.[3]
The DBD has straightforward function, the binding of DNA, which it accomplishes using mostly polar residues with some basic, cationic ones along the minor groove of the DNA.[4] Remarkably, the stabilization of the strand opposite the nick in a partially unwound state by the DBD facilitates ligation in trans.[5] The AdD serves as the catalytic core of DNA ligase I and operates via a highly conserved Lys residue.[6] The OBD actually has multiple conformations depending on whether or not DNA is bound.[7] Upon DNA binding, the OBD - Phe 635 and 872 sit in the minor groove and perhaps π-stack while Asp 570 and Arg 871 form salt-bridges connecting the AdD and OBD.[8] Unsurprisingly, most of these DNA binding interactions appear to be facilitated by metals.
The remaining N-terminus domain of DNA ligase I is involved in regulating interactions with outside proteins. Proliferating cell nuclear antigen (PCNA), DNA polymerase β (POLB), and replication factor C (RFC) are known to bind in to this domain of DNA ligase I.[9] PCNA, in fact, appears to lock DNA ligase into its catalytic state thereby increasing throughput.[10] The N-terminus interactive domain also has three Ser which can be phosphorylated; the phosphorylation of these residues at various parts of the cell cycle suggest regulation by post-translational modification.[11]
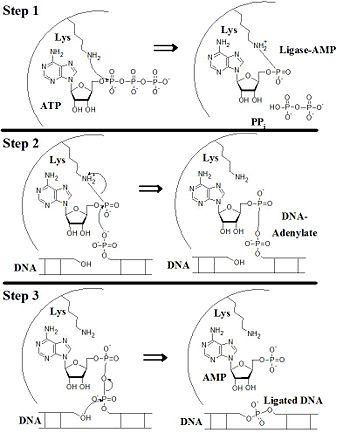
Ligating Mechanism
The process of ligation appears to occur in three steps, as shown in the figure. occurs when DNA is not bound and the OdD is flipped: Lys 568 nucleophilically attacks the α-P of AMP.[12] Unlike the scene shown for Step 1 (structure of this confirmation unavailable), the Arg 871 and Phe 872 that help to hold the DNA for ligation are exterior to the AdD. Instead, residues which flip to the outside of the protein upon DNA binding help to bind ATP.[13] The processes in Step 1 do form pyrophosphate that is not seen in the DNA-bound state shown fitting this model of conformational switching.
transfers the AMP from Lys 568 to the 5'-phosphate of the nicked DNA.[14] The Lys remains in close proximity probably stabilizing the large negative charge from the pyrophosphate intermediate. This exact intermediate was not crystallized; however, a 180° turn places the phosphates adequately close for bonding. Given the shifts in conformation observed in the OBD, this suggested shift in position from an unstable reaction intermediate seems reasonable.
accomplishes ligation of the nicked DNA. The 3'-OH of the other DNA strand attacks the pyrophosphate created in Step 2 producing double-stranded DNA and AMP.[15] Note the the AMP shown is lacking an oxygen which is illustrated as being on the 3'-C of what is a 2',3'-dideoxyribose - the presence of which prevented the ligation of the nicked DNA. Once ligated, the DNA and AMP leave the ligase active site, the OBD inverts to catalyze the Lys-ATP reaction, and further DNA ligations can occur.
Summary
DNA ligase I is a large, monomeric protein which ligates partial DNA strands resulting from Okazaki fragment replication of DNA. Apparently regulated by the binding of various other proteins and/or the phosphorylation of Ser residues in its N-terminus, the three other domains (DBD, AdD, and OBD) are required for ligation activity. Ligation proceeds with the OBD in one conformation enabling the formation of Lys-AMP from Lys 568 and ATP. Upon binding of DNA, the OBD is inverted helping the DBD and AdD hold the nicked DNA in a partially unwound state. From here, the 5'-end of the nick takes the AMP from the protein thereby forming a pyrophosphate. The 3'-OH then attacks, ligating the DNA and freeing AMP - all of which unbind to restart the process. As part of a larger family of ligases that are involved in replication like DNA ligase I, repair mechanisms, and recombination of DNA during meiosis, this family of proteins are fundamental to the operation of eukaryotic and prokaryotic cells alike.
References
- ↑ Ellenberger, T.; Tomkinson, A.E. Annu. Rev. Biochem. 2008, 77, 13-38; Shuman, S. J. Biol. Chem. 2009, 284, 17365-17369.
- ↑ Ellenberger, T.; Tomkinson, A.E. Annu. Rev. Biochem. 2008, 77, 13-38; Shuman, S. J. Biol. Chem. 2009, 284, 17365-17369; Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669.
- ↑ Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669.
- ↑ Ellenberger, T.; Tomkinson, A.E. Annu. Rev. Biochem. 2008, 77, 13-38.
- ↑ Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Ellenberger, T.; Tomkinson, A.E. Annu. Rev. Biochem. 2008, 77, 13-38; Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669;Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Pascal, J.M.; Tsodikov, O.V.; Hura, G.L.; Song, W.; Cotner, E.A.; Classen, S.; Tomkinson, A.E.; Tainer, J.A.; Ellenberger, T. Mol. Cell 2006, 24, 279-291
- ↑ Ellenberger, T.; Tomkinson, A.E. Annu. Rev. Biochem. 2008, 77, 13-38.
- ↑ Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669;Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669;Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.
- ↑ Tomkinson, A.E.; Vijayakumar, S.; Pascal, J.M.; Ellenberger, T. Chem. Rev. 2006, 106, 687-669;Pascal, J.M.; O'Brien, P.J.; Tomkinson, A.E.; Ellenberger, T. Nature 2004, 432, 473-478.