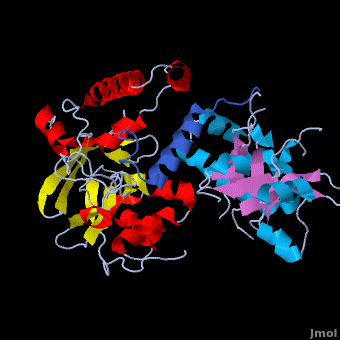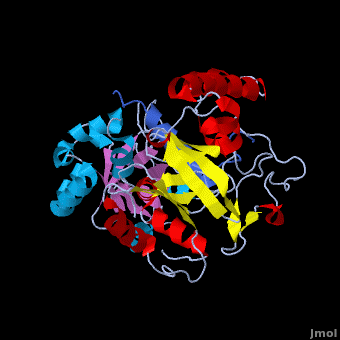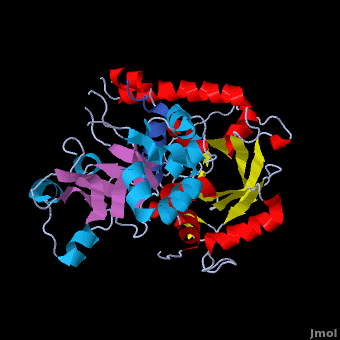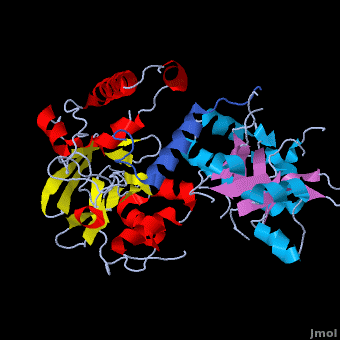
Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners
Orly Dym, Shira Albeck, Tamar Unger, Jossef Jacobovitch, Anna Branzburg, Yigal Michael, Daphna Frenkiel-Krispin, Sharon Grayer Wolf, and Michael Elbaum [1]
The soil bacterium Agrobacterium tumefaciens is a natural pathogen with the unique ability to genetically transform plants resulting in the Crown Gall disease. In addition to DNA, the bacterium delivers an abundant ssDNA binding protein, VirE2, whose roles in the host include protection from cytoplasmic nucleases and adaptation for nuclear import. , is presented here in complex with . VirE2 exhibits , each of which presents a combination with a . This combination defines the novel VirE2 fold. The structural similarity between the two domains is remarkable (RMSD of 3.1Å) considering the very low sequence homology between them (14.3%). Also notable is that are connected by a single, which exhibits no electron density for residues 343 and 344 suggesting that it is flexible. In , the two folded domains of . tightly around the single alpha-helix of VirE1. Both and hydrophobic interactions with VirE1 cement the two domains of VirE2 into a “locked” conformation where the flexible extended linker joining these two independent VirE2 domains does not constrain their relative orientation. Most of the interactions are electrostatic, involving between residues R168, K248, H315, R367 and K471 from VirE2 and N34, D40, E42, E45, E47 and N48 from VirE1. The to its strong electronegative surface resembling that of ssDNA. VirE2 can bind alternatively to VirE1 or to ssDNA. The acidic nature common to both substrates suggests that they bind via electrostatic interactions to a common region of VirE2. The also contain clusters of aromatic residues with side chains pointing toward the VirE1-binding site forming hydrophobic pocket. include Y160, Y194, Y373, Y319, W323 and W383, with the last three being involved in VirE1 interactions. These planar side chains may form additional stacking interactions with the nucleotide bases of DNA. This analysis does not rule out additional interactions between ssDNA and parts of VirE2 that do not interact with VirE1. It appears that the two domains of VirE2 acquire a different relative orientation upon binding of VirE1 versus ssDNA utilizing conformational changes enabled by the extended flexible linker. In the presence of ssDNA, VirE2 adopts a solenoidal form whose three-dimensional structure has been characterized previously using electron microscopy (EM) and a real-space helical image processing approach[2]. A comparison of the crystal structure of VirE2 in complex with VirE1, to the EM reconstruction of VirE2 in the presence of ssDNA, supports rearrangement of the VirE2 domains. Thus VirE2 appears as a dynamic structure that can accommodate its different partners due to the flexibility of its inter-domain linker. These structural features reflect the physiological requirements of VirE1 as an inhibitory chaperone of VirE2 in the bacterium, and of VirE2 as a chaperone of the ssDNA in the plant.