Sandbox Reserved 452
From Proteopedia
Contents |
C-Myc
Introduction
C-Myc is a transcription factor involved in the cell cycle specifically with the stimulation of cell proliferation but is also involved in the regulation of apoptosis, cell differentiation, cell competition, and senescence [1]. C-Myc is a DNA binding protein and functions primarily by inducing transcription through the formation of heterodimers primarily with Max proteins. C-Myc's interactions are not limited to only Max protiens as it has been shown to interact with BRCA1[2], MAPK1[3], p73[4], and many more. C-Myc is also a protooncogene, and when normally expressed is strictly regulated by signals within the cell cycle, so that when a cell is resting its c-myc expression is little while when growing it expresses high levels of c-myc. When normal cells have overexpression of c-myc they activate a protection pathway, the abnormal cells are thus eliminated from the host organism through apoptosis, thus protecting the organism.
Structure
Secondary Structures
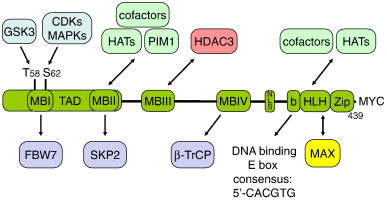
The c-myc protein contains two specific regions that characterize them as transcription factors: a carboxy-terminal basix helix-loop-helix leucine zipper (bHLH-LZ) motif and an amino terminal transactivation domain[5]. The N-terminal domain functions in the activation of transcription, and the C-terminal, which contains the bHLH-LZ functions in in promoting the interaction between of c-myc and max. Immediatley following the basic region within the c-myc protein is the helix -loop-helix motif which has been shown to interact with DNA through binding [5]. C-myc by itself contains 3 alpha helices and 74 residues. Since c-myc is essentially inactive when it homodimerizes and has a very short half life, typically 20 to 30 minutes, it heterodimerizes with max to perform its biological functions.
Active Site
The of c-myc is where it binds to the DNA after it has formed a heterodimer with max.
The figure below shows the structural organization of c-myc the bHLHZip hetereodimerizes with max and then they bind to DNA[6]
|
Circular Dichromism and NMR
In order to find more about the transciptional and biological activity of c-myc more about it's structure and heterodimerization had to be known. The two main techinques used to identify the bHLH-LZ domains of c-myc were Circular Dichromism(CD) and Nuclear Magnetic Resonance(NMR). The CD spectrum was used to compare v-myc with c-myc to identify alpha helical characteristics of the protein. It was also revealed through the CD that the c-myc/max heterodimer is more stable than the max/mad heterodimer. Electrophoretic Mobility shift assays(EMSA) were use to determine c-myc's function in binding DNA. NMR was used to determine the hydrophobic core formed upon formation of the heterodimeric Myc-Max protein complex the functions in binding DNA[7].
Mechanism
Most of the biological functions that c-myc performs involves herterodimerization with Max. C-Myc primary function is activating the transcription of genes involved in cell proliferation, growth, and metabolism by binding to the DNA sequence CAGTG, also known as the Enhancer box, when it has dimerized with Max. Many studies have shown that without dimerizing to Max, c-Myc would not be able to bind to DNA, which makes their interaction important to cellular function[8][9]. When c-Myc/Max form a herterodimer and bind DNA transcription becomes activated. When Myc-Max are bound, this allows for proteins involved in transcription to bind like TBP, TBIIE, TRRAP, etc. In a normal cell c-myc is very regulated and expresses little c-myc when the cell is resting. When cells becomes stimulated by growth factors c-myc is drastically increased. Over expression of c-myc in a normal cells results in apoptosis so that the organism is protected from abnormal cell growth. In abnormal cell when the c-myc gene is over expressed leads to cancer.
Medical Uses
The over expression of c-myc is one of the most common findings in human cancer. However, it is still unclear how it acts to promote abnormal cell growth. Studies have shown that the greater amounts of c-myc present, the higher your chances are for having cancer. Researchers have been looking into the binding of c-myc with max to see if it would deregulate c-myc's expression. This takes the c-myc protein out of the organism all together. They also have looked at mutating certain regions where c-myc binds DNA to try and understand why it effects proliferation that eventually causes cancer. One of the most common forms of cancer that c-myc is involved in is Burkitts's lymphoma. In this cancer, c-myc proto-oncogene is moved from it's normal position on chromosome 8 by reciprocal translocation to an enhancer region close to chromosome 14, the gene that is capable of making antibodies. Since antibodies are frequently used in the body to maintain a healthy immune system, c-myc is frequently transcribed in this region. So c-myc ends up becoming uncontrollably transcribed when it is translocated to this region.
C-myc could potentially become useful in cancer therapy as a drug, but until more is known about its mechanism and binding, the cure for cancer will still be unknown.
References
- ↑ Uribesalgo I, Benitah SA, Di Croce L. From oncogene to tumor suppressor: The dual role of Myc in leukemia. Cell Cycle. 2012 May 1;11(9). PMID:22510570
- ↑ Wang Q, Zhang H, Kajino K, Greene MI. BRCA1 binds c-Myc and inhibits its transcriptional and transforming activity in cells. Oncogene. 1998 Oct 15;17(15):1939-48. PMID:9788437 doi:10.1038/sj.onc.1202403
- ↑ Gupta S, Davis RJ. MAP kinase binds to the NH2-terminal activation domain of c-Myc. FEBS Lett. 1994 Oct 24;353(3):281-5. PMID:7957875
- ↑ Watanabe K, Ozaki T, Nakagawa T, Miyazaki K, Takahashi M, Hosoda M, Hayashi S, Todo S, Nakagawara A. Physical interaction of p73 with c-Myc and MM1, a c-Myc-binding protein, and modulation of the p73 function. J Biol Chem. 2002 Apr 26;277(17):15113-23. Epub 2002 Feb 13. PMID:11844794 doi:10.1074/jbc.M111281200
- ↑ 5.0 5.1 Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994 Feb;4(1):102-8. PMID:8193530
- ↑ Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012 Feb 25;494(2):145-60. Epub 2011 Dec 28. PMID:22227497 doi:10.1016/j.gene.2011.12.027
- ↑ Fieber W, Schneider ML, Matt T, Krautler B, Konrat R, Bister K. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J Mol Biol. 2001 Apr 13;307(5):1395-410. PMID:11292350 doi:10.1006/jmbi.2001.4537
- ↑ Littlewood TD, Amati B, Land H, Evan GI. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992 Sep;7(9):1783-92. PMID:1501888
- ↑ Muhle-Goll C, Nilges M, Pastore A. The leucine zippers of the HLH-LZ proteins Max and c-Myc preferentially form heterodimers. Biochemistry. 1995 Oct 17;34(41):13554-64. PMID:7577944