Image:Replication fork.jpg
From Proteopedia
No higher resolution available.
Replication_fork.jpg (600 × 420 pixel, file size: 52 KB, MIME type: image/jpeg)
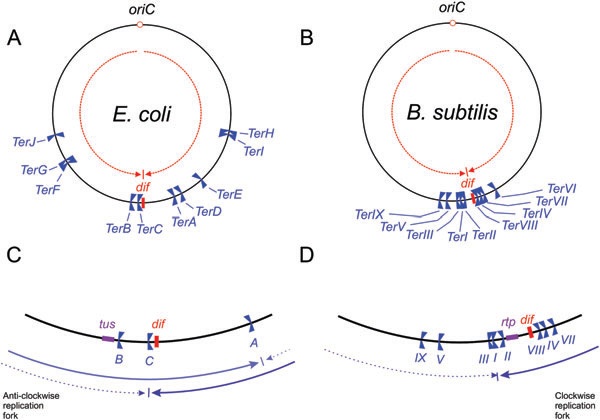
The DNA replication fork traps of the E. coli and B. subtilis chromosomes. A. The locations of Ter sites for E. coli (TerA–TerJ) are shown, as is the dif site. The two opposed groups of polar Ter sites form the fork trap; TerC, TerB, TerF, TerG and TerJ are orientated to block only clockwise moving forks, whereas TerA, TerD, TerE, TerI and TerH are orientated to block only anticlockwise forks. The dashed arrows indicate that DNA strand compositional skew organizes the chromosome into two segments bisected by the oriC and dif loci (see text). B. In the B. subtilis fork trap, TerI, TerIII, TerV and TerIX are orientated to block clockwise moving forks, whereas TerII, TerVIII, TerIV, TerVII and TerVI are orientated to block anticlockwise forks. C. The inner region of the replication fork trap of E. coli, showing the innermost Ter sites, the position of dif and the location of the tus gene. The continuous arrows indicate the movement of the first fork to arrive from oriC when the innermost Ter site blocks its movement (TerA or TerC, depending on whether the anticlockwise or clockwise fork arrives first). The dashed arrows indicate the movement of the second fork to arrive in each case. Thus, termination would occur at TerA if the clockwise replication fork were delayed, and at TerC if the anticlockwise fork were delayed. This would occur with reasonable frequency (Breier et al., 2005), but a significant number of forks would meet and fuse within the region between TerA and TerC. D. In B. subtilis, the Ter sites are more clustered towards the centre of the terminus region. Note that TerI and TerII are only separated by ~0.1 kb and the location of these two sites is not shown to scale. In B. subtilis 168, DNA replication almost always terminates at TerI because of the asymmetric location of TerI – the anticlockwise fork has to travel a longer distance than the clockwise fork to reach it. In other strains of B. subtilis that are cured of the SPb prophage on the anticlockwise replichore, the dif locus would be much closer to 180° from oriC. The tus (E. coli ) and rtp (B. subtilis) genes are autoregulated by the binding of the terminator proteins to Ter sites within the promoter regions (Ahn et al., 1993; Natarajan et al., 1991; Roecklein et al., 1991).
Source: Duggin et al. (2008)
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | User | Dimensions | File size | Comment | |
|---|---|---|---|---|---|
| (current) | 15:42, 22 May 2011 | Lauren Fowler (Talk | contribs) | 600×420 | 52 KB | The DNA replication fork traps of the E. coli and B. subtilis chromosomes. A. The locations of Ter sites for E. coli (TerA–TerJ) are shown, as is the dif site. The two opposed groups of polar Ter sites form the fork trap; TerC, TerB, TerF, TerG and TerJ |
- Edit this file using an external application
See the setup instructions for more information.
Links
The following pages link to this file:
