Ann Taylor p53 sandbox
From Proteopedia
p53 Tumor Suppressor Protein
p53, also known as TP53 (for Tumor Protein 53) was named "Molecule of the Year" by Science Magazine for its important role in both cell cycle regulation and apoptosis(). The name p53 reference to it apparent molecular mass. It runs as a 53 kDa molecule on SDS-PAGE. But based on calculations from its amino acid residues, p53's mass actually 43.7 kDa. This difference is due tothe high number of proline residues in the protein, which causes it to migrate slowly on SDS-PAGE.[1]
Human p53 is 393 amino acids long and has several domains:
1. The amino terminus (1-44) contains the transactivation domain which is responsible for activating downstream target genes.
2. A proline-rich domain (58-101) mediates the p53 response to DNA damage through apoptosis.
3. A core DNA-binding domain (102-292), which is the target of 90% of p53 mutations found in human cancers. A single mutation within this domain is sufficient to cause a major conformational change.
4. An oligomerization domain (325-256), which contains a β-strand which interacts with another p53 monomer to form a dimer, followed by an α-helix which mediates the dimerization of two p53 dimers to form a tetramer.
5. Two nuclear export signals (NES), found within the oligomerization domain, is rich in leucine residues and is highly conserved. It is proposed that oligomerization masks the NES, resulting in p53 remaining in the nucleus.
6. Three putative nuclear localization signals (NLS) in the C-terminus have been identified via mutagenesis and sequence similarity.
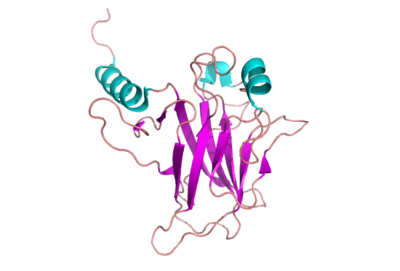
The intact p53 monomer is flexible and does not form orderly crystals. Consequently, the entire structure is not known. Instead, segments have been isolated and characterized. The figure at the right shows the cartoon representation of DNA binding domain that has been studied most rigorously.
|
p53 Pathway and mutation
In a normal cell p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage sensed p53's level rises(). p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the demage is repaired, or if the damage is unrepairable, p53 initiates the process programmed cell death,also known as apoptosis, permanently removing the damaged cell.()
In most of the human cancer p53 mutations has been observed. Most of the p53 mutations that cause cancer are found in and around the DNA binding surface of the protein.The most common mutation changes
that interacts with DNA when mutated to another amino acid this interaction is lost. Other residues that are commonly mutated are arginine 175, 249, 273, 282 and glycine 245.
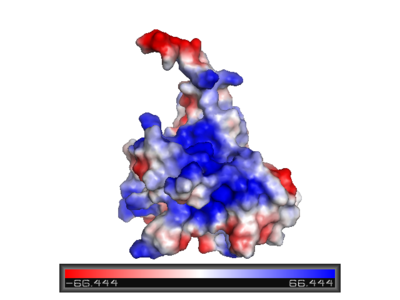
Surface charge of the DNA binding domain
The figure at the left shows the surface charge of the p53 DNA binding domain. It is rich in arginine amino acids to interact with DNA, thus this cause its surface positively charged. This domain recognizes specific regulatory sites on the DNA and flexible structure of p53 allow it to bind to many different variant of binding site allowing it to regulate transcription at many places in the genome.()
|
There is a Zn-binding motif on p53. The p53 Zn atom (shown in red) is coordinated by residues that are located on two loops, respectively. It is conceivable that the zinc plays a role of stabilizing two loops through coordination().