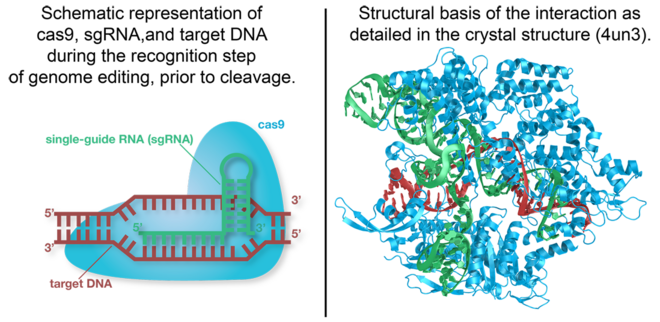
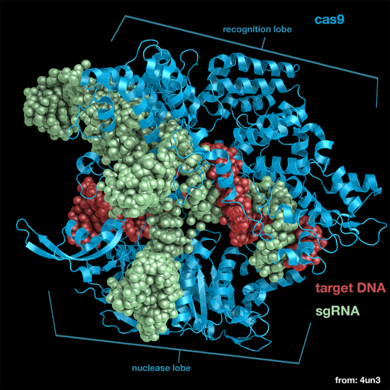
Cas9
From Proteopedia
Cas9 is the RNA-guided DNA endonuclease used by the CRISPR (clustered regularly interspaced short palindromic repeats)-associated systems to generate double-strand DNA breaks in the invading DNA during an adaptive bacterial immune response.
See also Cas9 (hebrew).
The CRISPR-associated endonuclease Cas9 has been exploited for use in genome editing systems. In such systems, an engineered single-guide RNA (sgRNA) is used to target double-stranded breaks in genomic DNA. Depending on what repair pathway is triggered, often dictated by the inclusion of additional engineered components, the targeted site either is disrupted or incorporates additional genetic sequences.
Microbiologist and 2020 Nobel laureate Emanuelle Charpentier, from Max Planck Institute for Infection Biology in Berlin, is one of the co-inventors of the groundbreaking
This video, by Paul Andersen, explains how the CRISPR/Cas immune system
was identified in bacteria and how the CRISPR/Cas9 system was developed to edit genomes.
a powerful new technology with many applications in biomedical research,
including the potential to treat human genetic disease.
Articles in Proteopedia concerning Cas9 include:
3D structures of Cas9
See Endonuclease 3D structures.
STRUCTURE OF Cas9 IN STAPHYLOCOCCUS AUREUS IN COMPLEX WITH sgRNA
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015 Aug 27;162(5):1113-26. doi: 10.1016/j.cell.2015.08.007. PMID:26317473 doi:http://dx.doi.org/10.1016/j.cell.2015.08.007
- ↑ Morlot C, Pernot L, Le Gouellec A, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae. J Biol Chem. 2005 Apr 22;280(16):15984-91. Epub 2004 Dec 13. PMID:15596446 doi:10.1074/jbc.M408446200
- ↑ Chen H, Choi J, Bailey S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J Biol Chem. 2014 May 9;289(19):13284-94. doi: 10.1074/jbc.M113.539726. Epub 2014, Mar 14. PMID:24634220 doi:http://dx.doi.org/10.1074/jbc.M113.539726
- ↑ Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014 Feb 27;156(5):935-49. doi: 10.1016/j.cell.2014.02.001. Epub 2014 Feb, 13. PMID:24529477 doi:http://dx.doi.org/10.1016/j.cell.2014.02.001
- ↑ Palermo G, Chen JS, Ricci CG, Rivalta I, Jinek M, Batista VS, Doudna JA, McCammon JA. Key role of the REC lobe during CRISPR-Cas9 activation by 'sensing', 'regulating', and 'locking' the catalytic HNH domain. Q Rev Biophys. 2018;51. doi: 10.1017/S0033583518000070. Epub 2018 Aug 3. PMID:30555184 doi:http://dx.doi.org/10.1017/S0033583518000070
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Michal Harel, Karsten Theis, Joel L. Sussman, Ann Taylor, Thomas Gastineau