Crystal structure of protein from Staphylococcus aureus
From Proteopedia
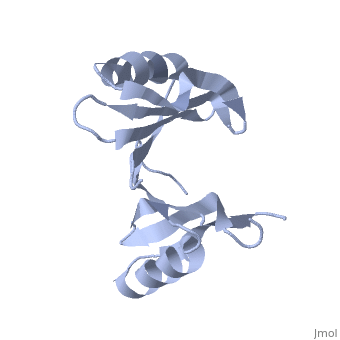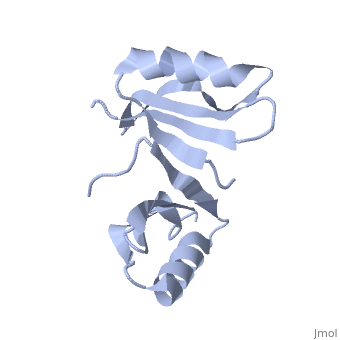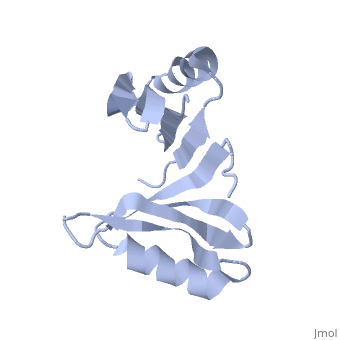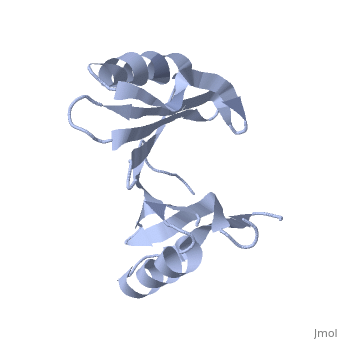
Crystal structure of hypothetical protein from Staphylococcus aureus (1tsj)PDB entry 1tsj refers to a hypothetical protein of 139 residues which is predicted as both a dimer[1] and a cytoplasmic protein.[2][3][4][5] The protein is associated with Pfam[6] entry PF06983 of 3-demethylubiquinone-9 3-methyltransferases. Among the sequence homologs found by PSI-Blast[7] there are the predicted 3-demethylubiquinone-9 3-methyltransferase proteins Q192X9 from Desulfitobacterium hafniense and A9VFW6 from Bacillus weihenstephanensis. Its potential substrate is S-adenosyl-L-methionine with the formal charge +1. PDB entry 2rk9 was found to be structurally[8] similar to 1tsj and share a significant sequence similarity[9] of 21.0%. This structure homolog is an Oxidoreductase from Vibrio Splendidus and binds methylglyoxal which is not charged. Another structural similarity was found with PDB entry 1t47 which is a 4-hydroxyphenylpyruvate dioxygenase, but the sequence similarity is lower (7.7%).
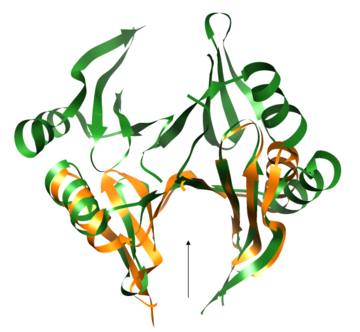
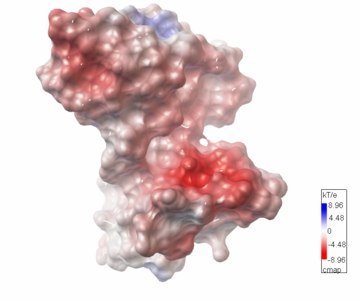
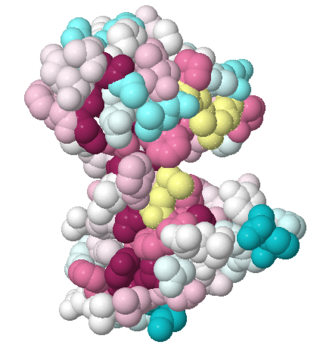
Both the sequence and structure homologs are from the same superfamily titled 'Glyoxalase/bleomycin resistance protein/dioxygenase'. The Superimposition[10] between chain A of 1tsj (orange) and chain B of 2rk9 (green) shows that the proteins indeed have similar folds (see Figure 1). It seems that the chosen active site of 1tsj is the largest cavity found on the protein's surface; in most cases this cavity is the functional area.[11] Evidence approving this choice is that the superimpositions with 2rk9 placed its known active site[12] at the same location. The electrostatic potential[13] on the protein's surface is mostly negative (see Figure 2). The evolutionary conservation[14] of 1tsj is shown in Figure 3.

It seems more likely that 1tsj belongs to the Glyoxalase/bleomycin resistance protein/dioxygenase superfamily and functions as a demethylubiquinone-9 3-methyltransferase protein than that it functions as Oxidoreductase.
|
| ||||||||||
References
- ↑ Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998 Sep;23(9):358-61. PMID:9787643
- ↑ Bhasin M, Garg A, Raghava GP. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 2005 May 15;21(10):2522-4. Epub 2005 Feb 4. PMID:15699023 doi:10.1093/bioinformatics/bti309
- ↑ Gardy JL, Spencer C, Wang K, Ester M, Tusnady GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FS. PSORT-B: Improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003 Jul 1;31(13):3613-7. PMID:12824378
- ↑ Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004 Mar 1;20(4):547-56. Epub 2004 Jan 22. PMID:14990451 doi:10.1093/bioinformatics/bth026
- ↑ Hua S, Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001 Aug;17(8):721-8. PMID:11524373
- ↑ Finn R, Griffiths-Jones S, Bateman A. Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics. 2003 May;Chapter 2:Unit 2.5. PMID:18428696 doi:10.1002/0471250953.bi0205s01
- ↑ Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997 Sep 1;25(17):3389-402. PMID:9254694
- ↑ Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The protein model portal. J Struct Funct Genomics. 2009 Mar;10(1):1-8. Epub 2008 Nov 27. PMID:19037750 doi:10.1007/s10969-008-9048-5
- ↑ Emmert DB, Stoehr PJ, Stoesser G, Cameron GN. The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res. 1994 Sep;22(17):3445-9. PMID:7937043
- ↑ Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605-12. PMID:15264254 doi:10.1002/jcc.20084
- ↑ Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998 Sep;7(9):1884-97. PMID:9761470
- ↑ Thornalley PJ. Glyoxalase I--structure, function and a critical role in the enzymatic defence against glycation. Biochem Soc Trans. 2003 Dec;31(Pt 6):1343-8. PMID:14641060 doi:10.1042/
- ↑ Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999 Feb;17(1):57-61. PMID:10660911
- ↑ Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009 Jan;37(Database issue):D323-7. Epub 2008 Oct 29. PMID:18971256 doi:http://dx.doi.org/10.1093/nar/gkn822
Created with the participation of Talya Etzion.
Categories: Staphylococcus aureus subsp. aureus | Burley, S K. | Gorman, J. | Min, T. | NYSGXRC, New York Structural GenomiX Research Consortium. | Shapiro, L. | Conserved hypothetical protein | Crystal structure | New york structural genomics consortium | New york structural genomix research consortium | Nysgxrc | Protein structure initiative | Psi | Structural genomic