Hox protein
From Proteopedia
This page, as it appeared on July 20, 2012, was featured in this article in the journal Biochemistry and Molecular Biology Education.
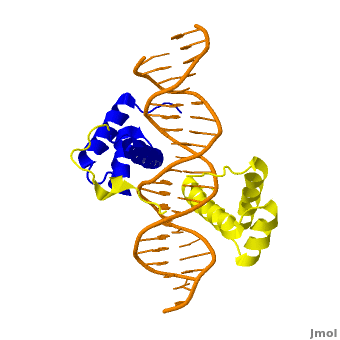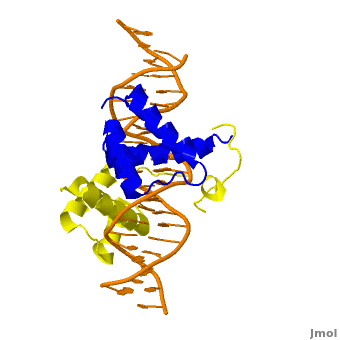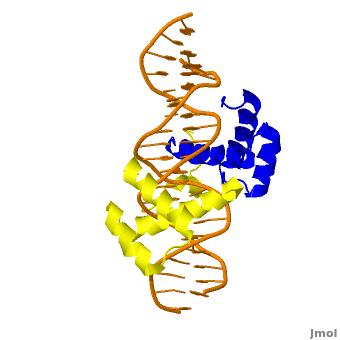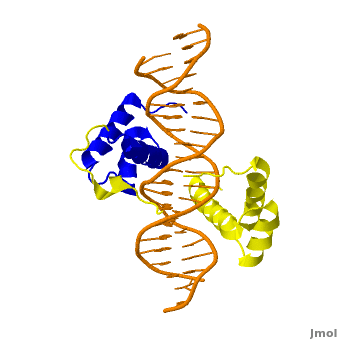
This is a joint project of students at La Cañada High School, La Cañada Flintridge, California USA, and students at the University of Southern California, Los Angeles, California USA, mentored by Professor Remo Rohs. Hox Proteins Recognize the Sequence-Dependent Shape of the Minor GrooveIntroduction and Biological Role of Hox Proteins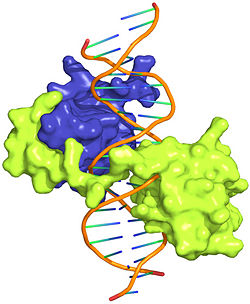 Figure 1: Crystal structure of Exd-Scr-DNA ternary complex; PDB ID# 2R5Z[1]. The Hox protein Scr (yellow) and its cofactor Exd (blue) bind to its specific fkh20 site. 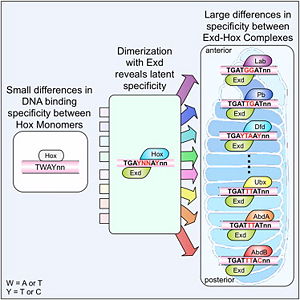 Figure 2: Hox proteins require a cofactor to achieve high binding specificity in order to execute their distinct functions in developing various parts of the fly embryo. Elsevier/Cell Press has provided permission for usage of this figure[2]. Hox proteins or homeobox proteins are transcription factors that play a key role in the embryonic development across species by activating and repressing genes. In Drosophila, eight Hox proteins are responsible for the development of different body segments of the fly, such as its antennae, wings, or legs. Hox proteins execute their distinct functions through binding to similar but different in vivo binding sites[3]. This page discusses molecular mechanisms through which Hox proteins recognize their DNA targets with very high binding specificity. The crystal structure of a Hox-DNA complex (Figure 1) shows that the Hox protein Sex combs reduced (Scr) binds its specific in vivo site with the help of cofactors, Extradenticle (Exd)/Pbx proteins. Hox proteins can bind DNA as monomers but their binding specificity is enhanced when the co-factor is present, a principle that is called latent specificity (Figure 2). In Drosophila, for instance, eight Hox proteins bind as heterodimers with their cofactor Exd to similar but distinct target sites. Hox proteins are expressed along the anterior-posterior axis of an embryo, thus determining the localization for the development of different body segments (Figure 2). This spatial order of expression from anterior to posterior is congruent with the location of the respective Hox genes at the chromosome, a fact known as collinearity. Hox mutants can lead to malformations, and studying the molecular basis of how Hox proteins execute distinct in vivo functions, therefore, remains an important field of biomedical research. Structural Description of Hox-DNA ComplexHomeodomain ArchitectureBoth Scr and Exd belong to the family of homeodomain proteins, which are encoded by homeoboxes. Homeodomains are helix-turn-helix motifs () comprised of three alpha helices (Figure 3). The DNA-binding interface residues of both proteins are . The third alpha helix of the Scr and Exd homeodomains, the so-called , inserts into the major groove where hydrogen bonds are formed between protein side chains and base pairs. An N-terminal tail forms contacts with the minor groove. Hox Protein-Cofactor Interactions Scr interacts with its cofactor Exd through hydrophobic interactions via a located at its N-terminal tail[4]. This interaction spans Scr's flexible N-terminal linker across the minor groove of its binding site. In the absence of the YPWM motif, Scr and Exd would not form a heterodimer. Major Groove Base ReadoutHox proteins achieve a large fraction of their binding specificity through . This form of protein-DNA recognition in the major groove is characterized as base readout since hydrogen bonds in the major groove can be used to distinguish between all four possible base pairs, A/T, T/A, C/G, and G/C. The Scr residues that engage in major groove base readout are Ile47, Gln50, Asn51, and Met54. Major groove contacts are almost identical across the Hox protein family and are not sufficient to achieve specificity within this family of transcription factors. Minor Groove Shape ReadoutMinor groove contacts, in addition to base readout in the major groove, provide the level of specificity that contributes to distinguishing factors within the Hox family. It has been shown that minor groove contacts are essential for achieving specificity[5]. Three side chains, of the Scr in vivo site fkh250. However, this additional level of binding specificity is not achieved through hydrogen bonds between protein side chains and functional groups of the bases. Such direct interactions are unable to distinguish A/T and T/A, or C/G and G/C base pairs due to the overlapping location of hydrogen bond donors and acceptors. The mechanism through which these three residues recognize the DNA minor groove is called shape readout as they do not form base-specific hydrogen bonds but rather recognize the sequence-specific narrowing of the minor groove. AT-rich regions can be characterized through an intrinsically narrow minor groove, leading to enhanced negative electrostatic potential, which in turn attracts basic side chains. This shape readout mechanism was found to be broadly employed by arginine residues [5]. Biological Function of Minor Groove Binding Residues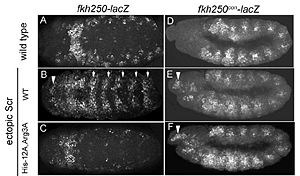 Figure 4: Expression patterns of Scr in presence of Scr specific site (left panel) vs. Hox consensus site (right panel). Elsevier/Cell Press has provided permission for usage of this figure[1]. In vitro binding studies have shown that His-12 and Arg-3 mutations have a large effect when exposed to the Scr specific site, whereas the effect is small when exposed to a Hox consensus site. The biological importance of both side chains becomes apparent in in vivo experiments. Upon mutations of His-12 and Arg-3 to alanine, Scr expression in a fly embryo is dramatically affected (Figure 4). In comparison to wild type Scr (A) and based on ectopic expression (B), there is only residual expression detected in
the thorax region of the double mutant when the Scr specific site is tested (C), whereas there is no apparent effect on expression in the presence of a Hox consensus site (D-F). Recognition of Scr Specific vs. Hox Consensus Site (PDB ID# 2R5Y)
This observation can be explained based on a second crystal structure of an Scr -Exd-DNA ternary complex where the Hox-Exd hetrodimer is bound to a Hox consensus site, which is not specific to Scr (). In this structure it is apparent that only and the remainder of the N-terminal linker is disordered (Figure 5). 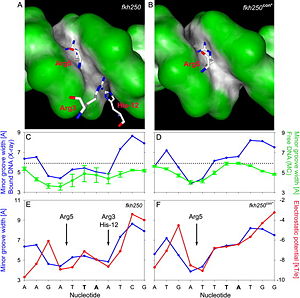 Figure 6: Comparison of DNA shape of Scr specific in vivo site (left panel) vs. Hox consensus site (right panel). Elsevier/Cell Press has provided permission for usage of this figure[1]. Based on the comparison of the two crystal structures of a Scr-Exd-DNA ternary complexes (Figure 6), it was found that three N-terminal residues contact the minor groove of the Scr specific site fkh250 (A) compared to only Arg5 binding the Hox consensus site fkh250con (B). In their protein-bound states, the shapes of both sites are distinct (dark gray, concave; green, convex surfaces). The distinct shapes of the two DNA binding sites, shown as minor groove width in the crystal structures of the complexes (blue plots), are already present when the protein is not bound to the DNA, with two minima in fkh250 (C) vs. one minimum in fkh250con (D), as inferred by Monte Carlo simulations (green plots). Minor groove width (blue plots) and electrostatic potential (red plots) correlate and form two binding pockets in fkh250 (E) and only a binding site for Arg5 in fkh250con (F). High-throughput Analysis of Hox-DNA Binding Specificity[[Image:Slattery-etal-Figure6.jpg |thumb|right|300px|Figure 7: DNA shape 3D structure of Hox proteinUpdated on 29-September-2021 1b72 – hHox-B1 + DNA – human 1puf – hHox-PRL + mHox-A9 + DNA References
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Remo Rohs, Eric Martz, Michal Harel, Joel L. Sussman, Skyler Saleebyan, Julia Tam, Bailey Holmes, Sharon Kim, Alexander Berchansky, Iris Dror, Ana Carolina Dantas Machado, Masha Karelina, Keziah Kim, Jaime Prilusky, Angel Herraez