Major Histocompatibility Complex Class I
From Proteopedia
Major Histocompatibility Complex (MHC) genes, and the proteins they specify, play centrally important roles in adaptive immune responses. Each MHC protein molecule contains a groove that is loaded with a peptide fragment derived from an intracellular protein. The MHC proteins carry these peptides to the outer surface of the cell, where thymus-derived ("T") lymphocytes examine them. When the peptides are deemed to be foreign by the T lymphocytes, appropriate immune defenses are activated. T lymphocytes are centrally important in all adaptive immune responses, including antibody production and the elimination of intracellular parasites, and their responses depend entirely on the presentation of peptides by MHC. Class I MHC proteins, in particular, reveal the presence of otherwise hidden intracellular parasites (such as viruses and some bacteria) by displaying peptide fragments of parasite proteins on the cell surface. For more detail, please see Wikipedia: Major Histocompatibility Complex.
The 3D structure of MHC proteins was one of the highest impact crystallographic strucures of all time. In order to appreciate why, some historical background is helpful.
Contents |
Major Histocompatibility Complex (MHC) Class I: Historical Background
Major Histocompatibility Complex (MHC) refers to a complex of closely linked genes first identified in the early to mid-20th century as being the major factors in the rejection of living tissue allografts (grafts between members of the same species). It was these studies that gave MHC its name. Many other genes contribute to tissue allograft rejection to minor degrees, and these were called minor histocompatibility genes. MHC genes code for MHC proteins that are the major antigens responsible for tissue allograft rejection. George D. Snell received one third of the 1980 Nobel Prize in Physiology or Medicine for his contributions to the identification and characterization of these genes. Of course many other researchers made crucial contributions and they are credited in Snell's Nobel Lecture. Jean Dausset received a third of the 1980 Nobel Prize in Physiology or Medicine for demonstrating the existence of MHC genes and proteins in humans, the latter being called Human Leukocyte Antigens (HLA). In mice, the most-used experimental model for studying MHC, the histocompatibility genetic loci were numbered H-1, H-2, H-3, and so forth. H-2 is the major histocompatibility locus, while all the others are minor. Both HLA and H-2 turned out to be large complexes of closely-linked genes.
Independently, in the early 1960's, Baruj Benacerraf and coworkers demonstrated the existence of immune response genes (Ir genes) that controlled the ability of an individual guinea pig's immune system to respond to simple synthetic amino acid polymers. Benacerraf was awarded one third of the 1980 Nobel Prize in Physiology or Medicine for discovering immune response genes. In the late 1960's, McDevitt and others found that the Ir genes were linked to MHC (for details, see Benacerraf's Nobel Lecture).
In 1975, Zinkernagel and Doherty made the surprising discovery that the ability of virus-specific T lymphocytes to recognize virus infection, in virus-infected cells, depended upon the MHC genotype of the infected cells. The MHC had to match that present when the T lymphocytes were first activated by the virus. This "restriction" of antigen recognition by T cells was confirmed in many other systems. In 1996, Zinkernagel and Doherty were awarded the Nobel Prize in Physiology or Medicine for this discovery. By that time, the mechanism of the "restriction" they had observed was clear -- thanks to the 3D structure of MHC Class I.
3D Structure and Its Significance
By the mid-1980's, there was abundant evidence that the ability of T lymphocytes to recognize antigen is "restricted" by MHC. However, what this "restriction" meant in terms of molecular mechanism was far from clear. Speculation about possible mechanisms raged for over a decade following Zinkernagel and Doherty's 1975 insight. But no experimental evidence available at the time was able to explain the "restriction". As an illustration, Figure 7 in Benacerraf's Nobel Lecture shows his thinking in 1980. The figure shows an "Ia molecule" hypothetically "specifically interacting" with an "antigen fragment". Note that although the genetic linkage between Ia (the molecule coded for by immune response genes) and MHC was well established, it was not yet clear that Ia was MHC. Benacerraf's thinking was correct, as far as it went, but the details were not yet available.
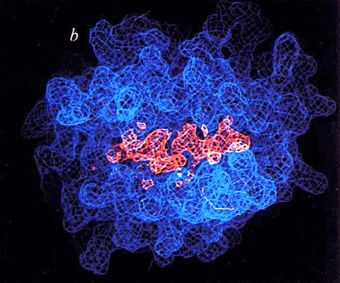
In 1987, Bjorkman and coworkers (in the laboratory of Don Wiley at Harvard) published the first empirical structure of MHC, a crystallographic structure of the human MHC Class I protein HLA-A2 (1hla). Although the resolution was low (3.5 Å), there was sufficient information to explain the decade-long mystery of how MHC restricts the recognition of foreign antigens by T lymphocytes. It is difficult to exaggerate the impact that this structure, and those that followed, had on the field of immunology.
Wiley's team struggled for many years to obtain sufficient MHC protein and high quality crystals. During this struggle, their funding ran out but they persevered, using funds from other projects (personal communication to Eric Martz from Wiley, ca. 1989). The MHC protein was obtained from cultures of human cells (the JY B lymphocyte cell line) by a well-established but arduous process that earlier had been used for obtaining the amino acid sequences of HLA proteins by Strominger and coworkers. Papain was used to cleave the soluble extracellular domains of the HLA MHC proteins from their transmembrane domains, and the HLA-A2 domains were purified, separating them from HLA-B7 among many other proteins present on these cells.
Because the protein was obtained from living cells, its peptide-accomodating groove (blue in figure at right) contained a mixture of unknown peptides (salmon-colored in figure at right). These appeared in the crystal structure of 1hla as an electron density that could not be explained by the known sequence of HLA-A2, lying within a groove the alpha chain of that molecule, in their Fig. 6b (at right). This phenomenally enlightening preview of ghostly peptides being presented to T cells by MHC gave goosebumps to cellular immunologists of the time.
MHC Structure Tutorial
A tutorial about the structure of MHC is available at MolviZ.Org. It includes side by side comparisons of two different viral epitopes in MHC class I (with synchronized mouse rotation), of epitopes in MHC I vs. II, and a chapter on MHC class II structure.
Recognition of MHC I (HLA A2:01) by T Cell Receptor Mimetic Antibodies
A recent study called Targeting a neoantigen derived from a common TP53 mutation describes the means by which T Cell Receptor mimic (TCRm) antibodies were created a new class of immunotherapy. TP53, tumor protein 53, is a tumor suppressor gene, and is the most commonly mutated protein in cancers. Most P53 is located inside the cell, the largest concentration is in the nucleus, which makes it difficult to find a treatment for TP53. Scientists have been struggling to design a drug to target this inactivated tumor suppressor gene. However, a small percentage is degraded by proteasomes and is present on the cell surface by MHC, Human Leukocyte Antigen (HLA). The most frequent mutation in the TP53 gene is a substitution from Arginine to Histidine at codon 175 (R175H). Most mutations within this gene occur as single-nucleotide variants at positions nearest to the DNA-binding domain. Using an HLA TP53 complex researchers discovered that neoantigen p53R175H is displayed by HLA:A2 on the cell surface at very low density. These peptide-HLA (pHLA) complexes are naturally ligands for T-cell receptors (TCRs). The advantage of using TCRm’s for immunotherapies is that they are easier to graft into different therapuetic formats and are an off the shelf ready to use therapy. Specifically, researchers developed a bispecific antibody constructed from H2 and an antiCD3 antibody, (H2- scDb) that can activate T cells even when the pHLA complex is expressed at very low, endogenous levels. This promising research shows that MHC I can be a key player in the fight against cancers caused by p53 mutations. [1]. See below for interactive figures from this research.
The video above depicts the 3D structure of the pHLA (p53R175H/HLA-A*02:01) that is bound to an H2-Fab fragment (PDB 6W51). The Fab fragment is colored according to its heavy chains (dark blue) and light chains (cyan). The Fab fragment is bound to the C terminus of the pHLA complex, where HLA-A*02:01 is colored gray and the β2 microglobulin is colored in gold. Sandwiched in between alpha helices of HLA are nine amino acids that are part of p53R175H shown light green.
This video shows the same structure from above, but zooming in shows the interaction of the pHLA with complementarity-determining regions. Hydrogen bonds are shown as dashed lines.
MHC Structures
A list of MHC structures is at Major histocompatibility complex.
References
- ↑ Hsiue EH, Wright KM, Douglass J, Hwang MS, Mog BJ, Pearlman AH, Paul S, DiNapoli SR, Konig MF, Wang Q, Schaefer A, Miller MS, Skora AD, Azurmendi PA, Murphy MB, Liu Q, Watson E, Li Y, Pardoll DM, Bettegowda C, Papadopoulos N, Kinzler KW, Vogelstein B, Gabelli SB, Zhou S. Targeting a neoantigen derived from a common TP53 mutation. Science. 2021 Mar 5;371(6533). pii: science.abc8697. doi:, 10.1126/science.abc8697. Epub 2021 Mar 1. PMID:33649166 doi:http://dx.doi.org/10.1126/science.abc8697
Proteopedia Page Contributors and Editors (what is this?)
Hannah Campbell, Eric Martz, Jaime Prilusky, Sandra B. Gabelli, Michal Harel
