MtrF
From Proteopedia
MtrF is a cell surface cytochrome on the Gram-negative bacteria known as Shewanella oneidensis.[1] MtrF is involved with shuttling electrons across its (S. oneidensis) outer surface. MtrF has several homologues, MtrC and the protein OmcA. These three different proteins are thought to be replaceable with one another in deletion mutation experiments.[1]
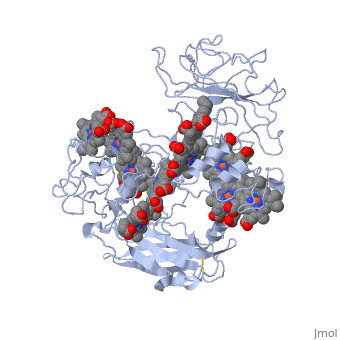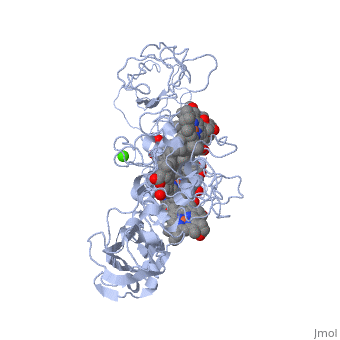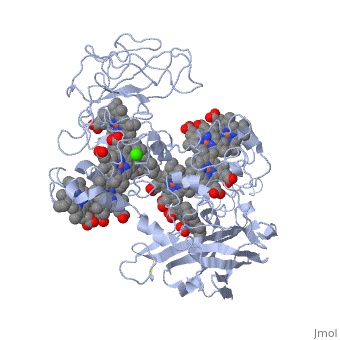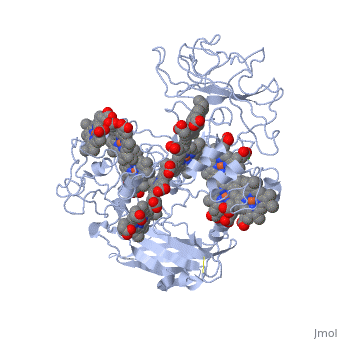
MtrF FunctionMtrF can play various roles in intermediating electron transfer straight to electron sinks, catalyzing electron exchange or partaking in extracellular intercytochrome electron exchange. Certain bacterial species (S. oneidensis) have the ability to utilize the extracellular mineral forms of iron and manganese as electron acceptors. In order for S. oneidensis to use these minerals, decaheme cytochromes must be present, they are positioned on the bacteria's cell wall at the endpoint of the trans-outer-membrane electron repositioning units. This process requires three different proteins to move electrons across the cell membrane; the process ends in a decaheme cytochrome labeled MtrF, in S. oneidensis. [1] MtrF StructureMtrF contains a few common elements; O,N,Fe and Ca. Oxygen is the most abundant element and is spread out through the entire protein. There are several nitrogens that form rings that have an iron placed in the center. There is also a lone calcium ion. There are alpha helices and beta sheets positioned throughout MtrF, there are more alpha helices but they are randomly placed in the structure while there are fewer beta sheets and they are located on opposite ends from one another. MtrF as mentioned above is a decaheme cytochrome, which means that there are ten heme groups that are spatially organized throughout the protein. Each heme is spread about 7Å from its neighbor, this close space allows for speedy electron transfer. The (ten) hemes are organized across four domains in a distinctive cross conformation. All of the hemes in the MtrF crystal structure display bis-His axial ligand coordination. The His residue in the domains provide the distal ligands for the five hemes in the same domain. Domains I and III contain antiparallel Beta-strands that connect two Greek key split Beta-barrel domains. Domains II and IV bind five closely packed hemes covalently attached Cys residues to the motifs in each domain. All the domains fold together so that the pentaheme domains II and IV are crowded together to form a central core with the two barrel domains I and III adjoining either side. The 3.2Å crystal structure proposes that the hemes, each corresponding to two His ligands, are near enough for effective electron transfer. Near-infrared magnetic circular dichroism and electron paramagnetic resonance spectroscopy provide additional support for these structural features. The complete structure of MtrF is similar to an ellipsoid with dimensions of 85x70x30Å. This particular structure was able to provide molecular insight into how the reduction of insoluble substrates, soluble substrates, and cytochrome redox partners may be possible together at different termini on an electron transport chain on the cell surface.[1] MtrF’s role in DiseasesNeisseria gonorrhoeae is the bacteria that causes the STD gonorrhea. This bacteria is quite resistant to many hydrophobic drugs, detergents, and dyes. The reason behind the tough resistance is the energy dependent efflux pump Mtr (Multiple transferable resistance). MtrF has been identified as a cell envelope protein that is involved with the resistance of hydrophobic antimicrobials in Neisseria gonorrhoeae. MtrF is a protein that helps highlight the expression of detergent resistance by Gonococci. MtrF is thought to act in accordance with the MtrC- MtrD- MtrE efflux pump; to make sure Gonococci has high level resistance to specific hydrophobic agents. MtrF is located near the MtrR gene and is predicted to encode a cytoplasmic membrane protein that contains up to twelve transmembrane domains. The expression of MtrF is ultimately subject to MtrR’s transcriptional control. MtrF was given its name because it is so closely tied to the protein MtrR. Several orthologues were discovered in a few Gram-negative and positive bacteria, indicating that perhaps the predicted products may represent an undescribed protein family that is highly involved with resistance of antimicrobials. [2] Regulation of MtrFA prime example of MtrF regulation is the study of "Regulation of MtrF Expression in Neisseria gonorrhoeae and Its Role in High-Level Antimicrobial Resistance".[3] In this study the expression of MtrF was repressed by MtrR (the major repressor in the mtrCDE expression). Another repressor known as MpeR can also regulate the expression of MtrF. Repression of MtrF by MtrR and MpeR was used, demonstrating that the repressive effects mediated by these regulators are independent processes. MtrF was also disabled and the significant reduction in the induction of hydrophobic agent resistance and it was found that the expression of MtrF is enhanced when Gonococci are grown under inducing conditions.
|
| |||||||||||
3D Structures of Cytochrome C
References
- ↑ 1.0 1.1 1.2 1.3 Fotinou C, Emsley P, Black I, Ando H, Ishida H, Kiso M, Sinha KA, Fairweather NF, Isaacs NW. The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin. J Biol Chem. 2001 Aug 24;276(34):32274-81. Epub 2001 Jun 19. PMID:11418600 doi:10.1074/jbc.M103285200
- ↑ Veal WL, Shafer WM. Identification of a cell envelope protein (MtrF) involved in hydrophobic antimicrobial resistance in Neisseria gonorrhoeae. J Antimicrob Chemother. 2003 Jan;51(1):27-37. PMID:12493784
- ↑ Folster JP, Shafer WM. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J Bacteriol. 2005 Jun;187(11):3713-20. PMID:15901695 doi:10.1128/JB.187.11.3713-3720.2005
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Holly Huntley, Alexander Berchansky