Sandbox Reserved 1481
From Proteopedia
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Crystal structure of the catalytic domains of Mettl3/Mettl14 complex
|
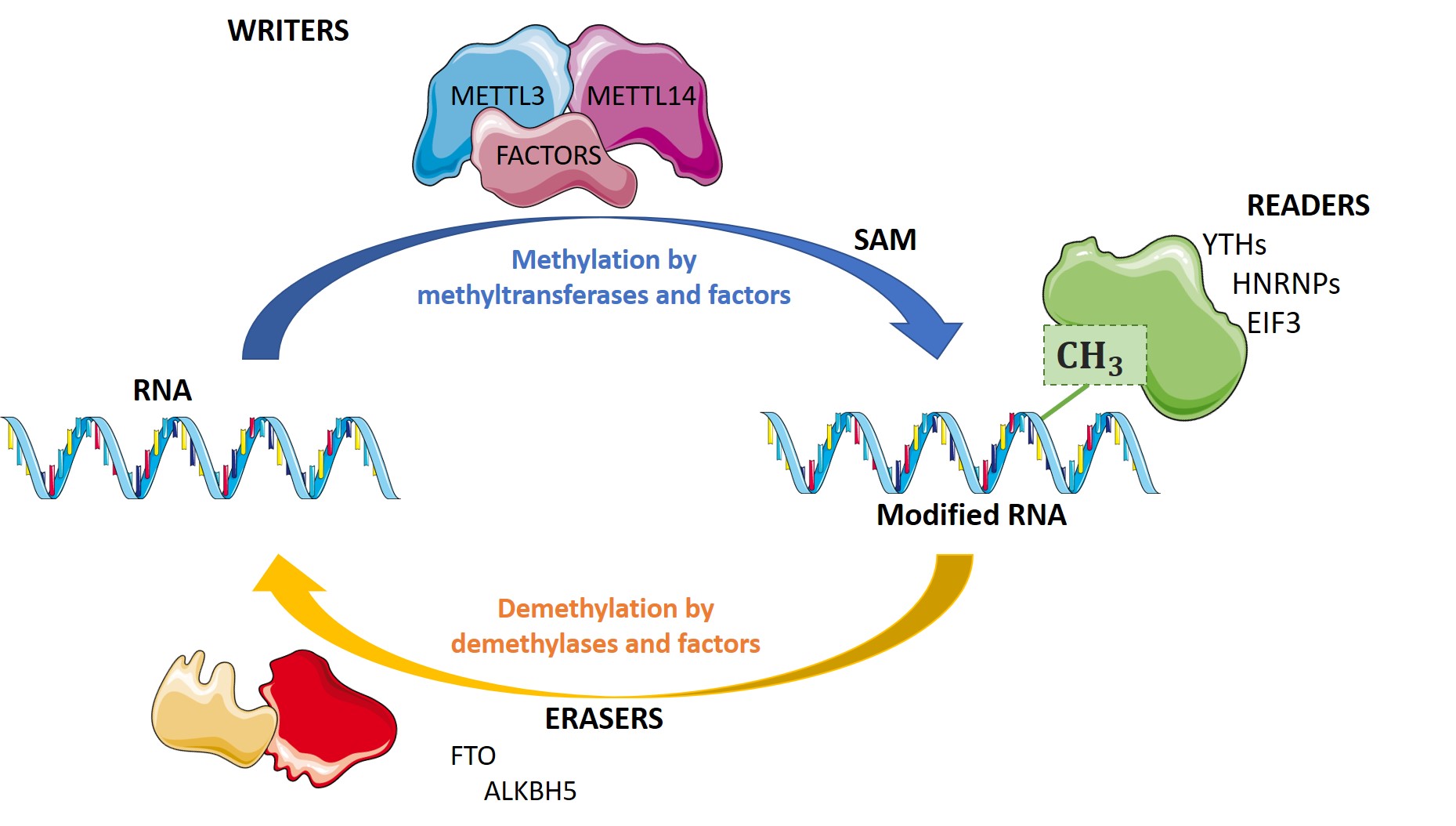
The complex METTL3/METTL14 is a heterodimer enzymatic complex involved into RNA post-transcriptional modifications by humans. This complex is abble to add a methyl group on adenosin of the RNA, by catalyzing a m6(A) modification.The N(6)-methyladenosine (m(6)A) is a quite common, reversible chemical modification of RNAs molecules, which plays a key role in several different biological functions. This post-transcriptional modification can be added by WRITERS, recognized by READERS and also removed by ERASERS. The METTL3/METTL14 complex plays the role of writer.
This enzymatic complex belongs to the second class of enzymes, which are the transferases and to the group of N6A-MTases. The complex is formed by 574 amino acid residues, divided into two different proteins nammed as Methyltransferase Like number 3 and 14.
Structural highlights of the METTL3/METTL14 complex
Primary structure
This complex is composed of two differents proteins forming two distinct but linked subunits of the enzymatic complex. Each of these two proteins is coded by one different gene. The Mettl3 gene codes for the N6-adenosine-methyltransferase 70 kDa subunit, which is a 225 Amino acid chain, whereas the Mettl14 gene codes for the N6-adenosine-methyltransferase subunit METTL14, which is composed of 349 amino acids. The complex is regulated by a supplementary subunit, the WTAP (Wilms' tumour 1-associating protein),that bind to METTL3/METTL14.
Each of these two proteins are corresponding to the A [2]or the B chain [3] of the complex and are neccessary to the functionning of the whole complex.
https://www.uniprot.org/uniprot/Q9HCE5
Secondary structure
Both polypeptides METTL3 and METTL14 are able to form some secondary structures thanks to interchain and intrachain hydrogen bonds formation. The core of METTL3 and METTL14 is shaped by Rossman fold domain,comprising a beta sheet edged by four alpha helices.
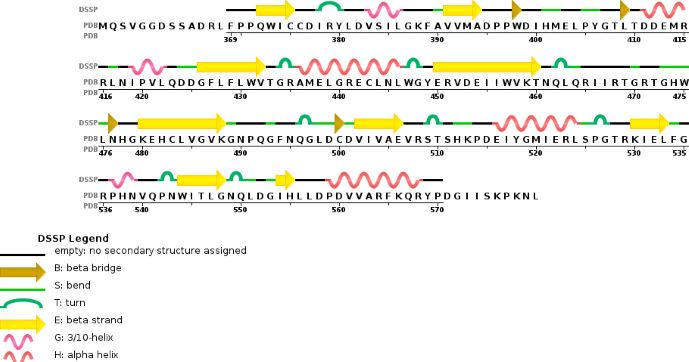
The A chain corresponding to METTL3, is mainly composed of 20% helical structures (7 helices; 46 residues) and 24% parallel and anti-parallel beta sheets (13 strands; 55 residues)
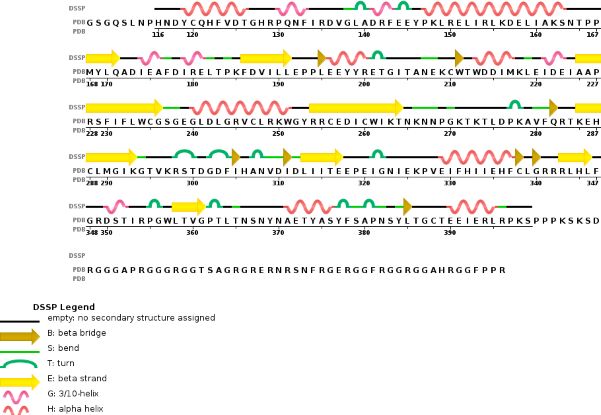
The B chain corresponding to METTL14 is made of 24% helical structures (14 helices; 87 residues) and 18% parallel and anti-parallel beta sheets (16 strands; 63 residues)
Tertiary structure Thanks to their primary amino acid sequences both A and B chains have some specific conserved domains directly linked to their function and functionning.
Domains and specific site or sequences
METTL3 and METTL14 are linked to each other by a an extensive water-mediated hydrogen bond, which generates a positively charged groove. This groove is well-conserved throughout evolution and is composed of four amino acids of each METTL3 and METTL14. The positive charge is due to the four amino acids of METTL14 which are arginins forming a pocket occupied by an acetate ion.
MTD : Methyltransferase domain
METTL3 and MTTL14 have both a methyltranferase domain, which is the domain able to catalyze a methyltransferase reaction. But the complex METTL3/METTL14 has a better methyltransferase activity, than one single subunit activity alone. Moreover, a mutation in the catalytic center of METTL3 inhibits the whole methyltransferase activity of the complex, whereas a mutation in the catalytic center of METTL14 does not. Thus, METTL3 is the catalytic subunit of the complex and METTL14 enhances the methyltransferasese activity. Indeed, it stabilizes the complex structure and binds to messenger RNA by recognizing its consensus sequence GGACU, which is the target for m6A modifications.
The MTD domain of METTL3 has an α–β–α sandwich fold. First, there are four α-helices : on one side there are α1,α2,α4 and on the other side there is α3. Then there is a mix of eight β-sheet with a specific order and direction : β1↑, β8↑, β7↑, β2↑, β3↑,β5↓, β4↑ and β6↑. There are three other α-helices to complete the fold.
Both MTD have approximatly 25% sequence homology. Despite some common points, the methyltransferase domain of METTL14 has extra terminal extension, with an unusual N-terminal extension which is approximately 50 amino acids long. On one hand, the N-terminal extension created along the helix, which goes through the domain, allows a close contact with the MTD of the catalytic subunit of the complex. This contact is made by several loops and shorter helical segments. On the other hand, the C-terminal helix of the MTD of METTL14 is antiparallel to the N-terminal extension helix, in order to stabilize its position. This allows the formation of a broad interdomain binding interface, and also other contact points along terminal extensions. These extra sequences are stabilizing the positions of both domains neccesary for the proper functionning of the enzyme.
|
Zinc finger binding domain of the METTL3 subunit, also called the CCCH sequence
Zinc finger domain of the METTL3, is one of the RNA binding domains of the complex. This perticular domain of the complex allows the METTL3 protein to make interactions with RNA molecule to modify via consensus sequences of the domain.
The CCCH sequence corresponds to a Cys-Cys-Cys-His motif. This specific motif belongs to METTL3 and belongs to the family of zinc binding motifs.
There are two of these CCCH motifs nearby the methyltransferase domain of METTL3. Zinc-binding motifs, like CCCHs are known for their abilities to bind nucleic acids such as RNA, their presence are required for RNA methylatransferase activity. In case of a punctual mutation in these sequences, a loss of the methyltransferase activity is observed, confirming their importance.
METTL3 and METTL14 are linked to each other to form the functionnal complex. So as the METTL3 is responsible of the catalytic activity, the METTL14 allows the allosteric recognition of messenger RNA. The two subunits' binding creates a cavity in the center of the complex where the enzymatic reaction takes place, it is the catalytic center. In this catalytic center some critical amino acid residues for the substrate recognition and specific binding are located. The residues D395, F429, Y406, W475, N477 on METTL3, the E192, R298,and D312 on METTL14 are all involved in binding of the RNA targeted sequence to the enzymes. These eight amino acids are considered as critica amino acids residues and their position in the 3D space in the catalytic center cavity is critical for the recognition of the enzyme's substrate. They are also playing a contact role with RNA's molecule to modify, and some of them are catalytic residues.
Crystal structure
To study the crystal structure of the METTL3/METTL14 some crystalograghy experiments have been realized under the following experimental conditions to crystalyze the protein complex:
Method : Vapor Diffusion Hanging Drop
pH 8
Temperature : 291.0 K
Buffer: 0.1 M Tris pH 8.0, 20% PEG3350
After crytalization, crystal had been analyzed thanks to X-Ray diffraction, at 100°C, with a single wavelength coming out of a synchrotron. Thus, the following data have been collected to describe the cristal structure of the complex :
Unit Cell caracteristics : Length (Å) a = 101.86 b = 101.86 c = 117.66
Angle (°) α = 90 β = 90 γ = 90
Symmetry : Space Group P 41 21 2
The primitive cubic symmetry of the unit cell is driven by two 2-fold axis rotation. Moreover, within this space group which is acentric, chiral, and enantiomorphic with one single lattice translation, eight differents representative symmetry operations are needed to get the whole unit cell.
The crystal structure analysis at 1,65 angstrom allows to collect precise and detailled informations about the whole structure like the side chains and also the hydrogen bonds. (Cristal analysis data at other resolution are available on [5].)
Complex METTL3/METTL14 enzymatic working process
The enzymatic complex of two proteins is able to realize methylation reactions thanks to its sequence and specific domains which allows it to catalyze some perticular reactions at given sites.
METTL3/METLL14 are belonging to the Writer's group, which is responsible of the realization of post transcriptional modifications of the RNA, within the humans cells. The METTL3/METTL14 complex catalyzes the methylation reaction which takes place in the nucleus.
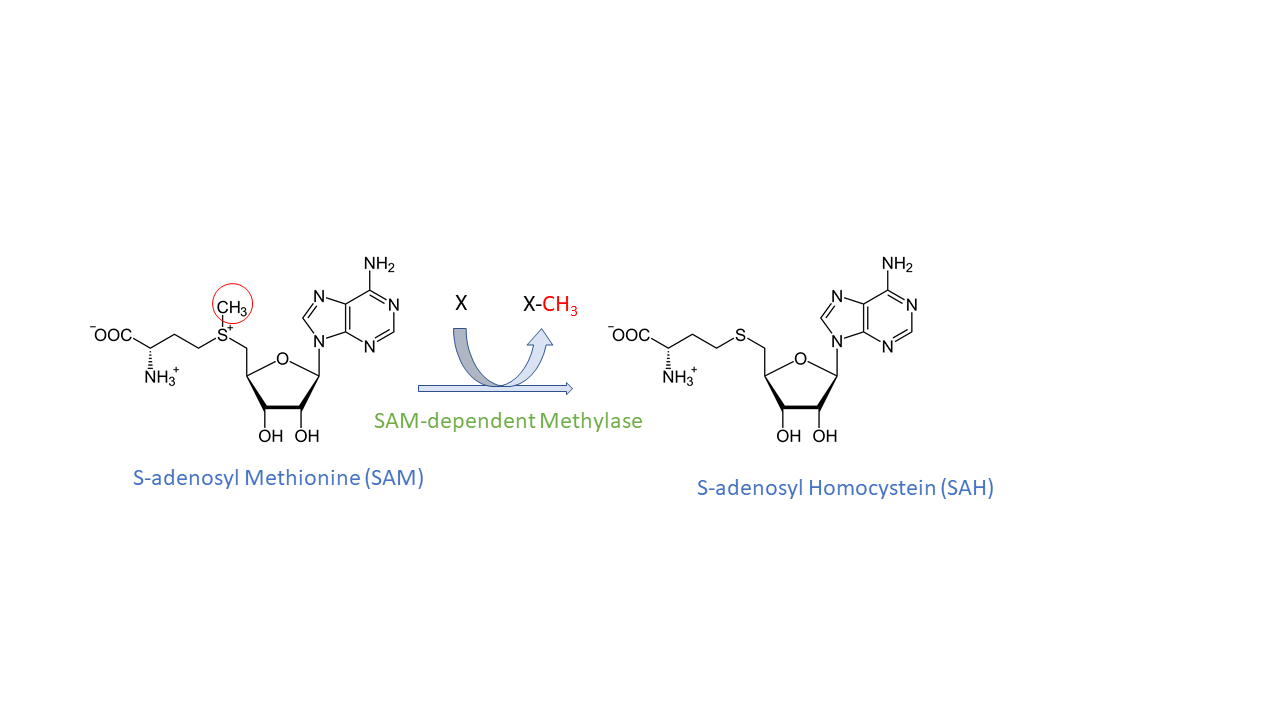
As previously said, the METTL3 is the catalytic component of the methyltransferase complex. So it is responsible of the m6A modifications thanks to its ability to crosslink with an other essential protein the S-adenosylmethionine (SAM). SAM is the substrate donor of the reaction. It is bound to a specific site of METTL3, at the C-ter of beta 1, beta 7, and beta 8 strands. Then, it is transformed into S-adenosylHomocystein (SAH). Moreover, METTL3 can be associated with METTL14, which contains its own methyltransferase domain. Even if the precise molecular interactions between the two subunits are unknown, it is known that both have to associated with each other, but also that aditionnal factors are neccessary to allow the addition of a methyl group on the targeted RNA.
After the formation of the minimal complex with the crystallized heterodimer of methyltransferase domains from METTL3 and METTL14 and two CCCH motifs of METTL3, and the binding of all the factors, the RNA modification reaction can take place. The acetate ion encasted in the arginin's pocket of the positively charged groove mediates RNA binding by mimicking RNA phosphate group.
Function of the modification within the cell
Specific, and well controlled methylation of the nucleic acids is essential for proper gene regulation, whatever the studied organism or modification’s type. However, the precise molecular role of m6A is unknown, it is known that for most mRNAs, m6A modifications appear in long exons, near stop codons, and in 3′ UTRs. This modification could influence mRNA stability, induce RNA conformational changes, modulate protein-RNA interactions, and even modify microRNA processing. One of the most prevalent modifications observed for mRNAs is N6-methyladenosine (m6A). Recent studies have intensely investigated how m6A-modification of RNA contributes to central events in biology. Nonfonctional m6A actors such as writers like METTL3 or METTL14, but also readers, and erasers are oftern linked to problems in self-renewal of stem cells, circadian clock and developmental defects, obesity, synaptic signaling, and cancers.
Disease and problems triggered by unfunctionnal METTL3/METTL14 complex
The m6A modification is a process involved in stem cells differenciation, mamalian circadian clock and embryonic development. Thus, an unfunctional METTL3/METTL14 complex that impacts m6A impacts these mechanisms.
Inhibition of METTL3
m6A stimulates the RNA export out of the nucleus and eases mRNA translation by recruiting EIF3, involved in the initation machinery of translation. Therefore, its inhibition could alter the translation process and lead to unfunctional proteins.
WTAP splicing factor
Wilms tumor 1-associated protein is an oncogenic protein, which regulates METTL3/METTL14 complex. It is known that its inhibition reduces the ability of METTL3 to bind mRNA. However, there are no evidence that its function of regulating the complex could contribute to its oncogenic role, according to "METTL3 regulates WTAP protein homeostasis", Melissa Sorci, Zaira Ianniello, Sonia Cruciani, Simone Larivera, Lavinia Ceci Ginistrelli, Ernestina Capuano, Marcella Marchioni, Francesco Fazi & Alessandro Fatica, Cell Death & Disease Article Published: 23 July 2018.
Stem cell differenciation
A knockdown of METTL3/METTL14 in embryonic stem cells induces a loss of m6A and an inability of self-renewal. The loss of m6A stops the embryonic stem cell to leave the pluripotent state for differenciation.
Cancer progression
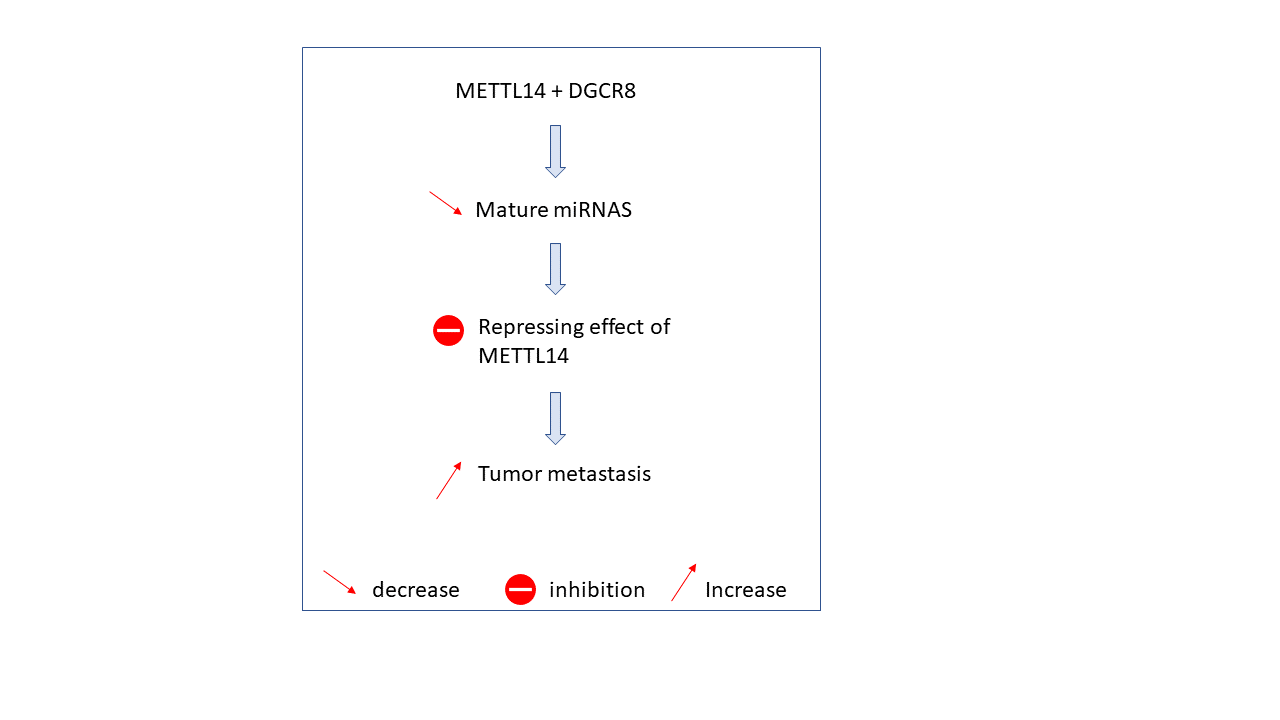
METTL3 plays a role in alternative pre-mRNA splicing. Indeed, a lack of METTL3 leads to a changement of the gene expression and alternative splicing related to the P53 signaling pathway wich is a tumor protein. m6A influences the processing of micro-RNAs and long non-coding RNAs splicing, which both are translational controller, and that can garble cancer progression. MeTTL3 is directly linked to lung cancer by promotting an increasing translation of mRNAs thanks to an interaction with translaion iniatiation factors (CBP80 and EIF4E). METTL14 has a repressing effect in tumor metastasis, but by interacting with DGCR8 (a protein required for micro-RNAs processing) instead of METTL3, it decreases mature micro-RNAs which at first instance inhibits the repressing effect of METTL14 in tumor metastasis, so it promotes growth of cancer cells and espacially Hepatocellular carcinoma.
Breast Cancer
It observed that in case of breast cancer, there is less and less METTL14 present in cells and is related to a shorter survival patients.
Glioblastoma
Glioblastoma stem cells' self-renewal is regulated by METTL3 thanks to its methyltransferase catalytic activity.Therefore, inhibiting all the METTL3/METTL14 complex or inhibiting only METTL3 or METTL14 leads to a reduced catalytic activity of METTL3 which is not able to regulate glioblastoma stem cells' self-renewal annd leads to the development of the brain tumor.
Endometrial cancer
Endometrail cancer patients often show to have a mutation: substitution mutation of an arginin to a proline. It is about an evolutionary conserved arginin (Arg 298) of METTL14 in the MTD3/MTD14 junction. This mutation leads to a deacrising methyl activity of the METTL3/METTL14 complex with the inability for it to recognize the RNA target.
Intestin cancer
There is a well-conserved trhoughout evolution tyrosin residue(Y406) located in the the active site of METTL3 that is required for METTL3 to interact with the RNA that needs to be methylated. Indeed, a mutation of this tyrosin is often found in intestin cancer patients.
References
[1] [2] [3] [4] [5] [6]/ [7] [8] [9] [10] [11] [12]
- ↑ Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016 Jul 21;63(2):306-17. doi: 10.1016/j.molcel.2016.05.041. Epub 2016 , Jun 30. PMID:27373337 doi:http://dx.doi.org/10.1016/j.molcel.2016.05.041
- ↑ Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016 May 25;534(7608):575-8. doi: 10.1038/nature18298. PMID:27281194 doi:http://dx.doi.org/10.1038/nature18298
- ↑ Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Corrigendum: Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2017 Feb 9;542(7640):260. doi: 10.1038/nature21073. Epub 2017 Jan 18. PMID:28099411 doi:http://dx.doi.org/10.1038/nature21073
- ↑ Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016 Sep 14;5. pii: e18434. doi: 10.7554/eLife.18434. PMID:27627798 doi:http://dx.doi.org/10.7554/eLife.18434
- ↑ Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: Two subunits, 2 roles. RNA Biol. 2017 Mar 4;14(3):300-304. doi: 10.1080/15476286.2017.1282025. Epub 2017, Jan 25. PMID:28121234 doi:http://dx.doi.org/10.1080/15476286.2017.1282025
- ↑ doi: https://dx.doi.org/10.2210/pdb5K7M/pdb
- ↑ Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015 Jul 1;29(13):1343-55. doi: 10.1101/gad.262766.115. PMID:26159994 doi:http://dx.doi.org/10.1101/gad.262766.115
- ↑ Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014 Feb;10(2):93-5. doi: 10.1038/nchembio.1432. Epub 2013 Dec 6. PMID:24316715 doi:http://dx.doi.org/10.1038/nchembio.1432
- ↑ Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018 Jan 26;9(2):124. doi: 10.1038/s41419-017-0129-x. PMID:29374143 doi:http://dx.doi.org/10.1038/s41419-017-0129-x
- ↑ Zhou KI, Pan T. Structures of the m(6)A Methyltransferase Complex: Two Subunits with Distinct but Coordinated Roles. Mol Cell. 2016 Jul 21;63(2):183-185. doi: 10.1016/j.molcel.2016.07.005. PMID:27447983 doi:http://dx.doi.org/10.1016/j.molcel.2016.07.005
- ↑ Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018 Jan 25;37(4):522-533. doi: 10.1038/onc.2017.351. Epub 2017 Oct 9. PMID:28991227 doi:http://dx.doi.org/10.1038/onc.2017.351
- ↑ Sorci M, Ianniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, Marchioni M, Fazi F, Fatica A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018 Jul 23;9(8):796. doi: 10.1038/s41419-018-0843-z. PMID:30038300 doi:http://dx.doi.org/10.1038/s41419-018-0843-z