Sandbox porins
From Proteopedia
This page is setup for Matt to build his senior project for OU CHEM 4923
|
Contents |
Porins Overview
Porins such as OmpC[1], are proteins located in the outer membrane of gram-negative bacteria or the outer membrane of mitochondria in eukaryotes and function as simple diffusion channels. The simple diffusion channels allow diffusion of sugars, ions, and amino acids to cross the outer membrane. Formation of porins occurs when a signal sequence is used to transfer the porin protein out from the inner membrane of bacteria. For this to occur, chaperone protein Sec B containing an unfolded polypeptide chain sends out a signal. This signal is sent to Sec A ATPase and binds to the Sec B complex. The polypeptide is then released and sent to the Sec E – Sec Y translocation channel where it begins to fold into partially assembled beta-sheets. The lipid-binding region located in the hydrophobic core of the partially assembled beta-sheets will then pull the complex into the outer membrane. Once in the outer membrane, the beta-sheets completely form by the alternating polar/non-polar side chains. Once porins have formed, structural components, functions, and energetic processes can be observed.
Structure
The general structure of porins is most recognized by its secondary structures.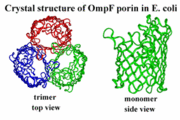
Function
Porins have many functions in context of its structural components. The main function of porins is passive diffusion channels so that sugars, ions, and amino acids may pass through the outer membrane. For this to occur however, several other functions from other components must occur. One of these components are the hydrophilic chains that line the inside of the beta-barrel of porins. The function of the hydrophilic chains is to create a polar environment within the ring to help sugars, ions, and amino acids to pass through. Another important component is the eyelet or constriction zone located inside the middle of the beta-barrel of porins. The eyelet blocks the water-filled channel of porins and determines what the size capacity and ion selectivity is for a solute to pass through. If a porin has salt-bridges between its N-terminus and C-terminus then it will create more stability for the structure. Lastly, the outside loops formed by the anti-parallel beta-sheets are rigid and packed close together. This allows porins to become highly resistant to proteases and detergents.
Energetics
The components and functions of porins can be brought together with its energetics to explain porins main function, simple diffusion.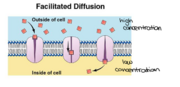
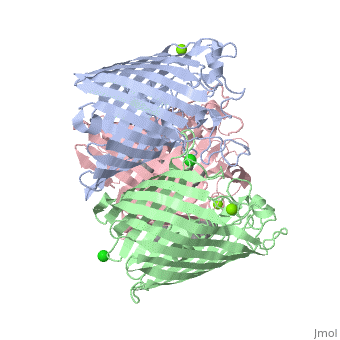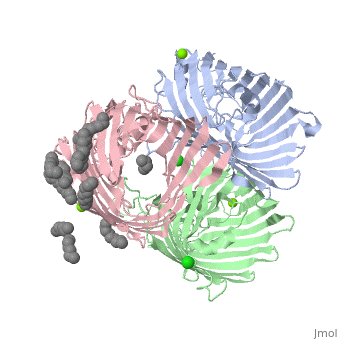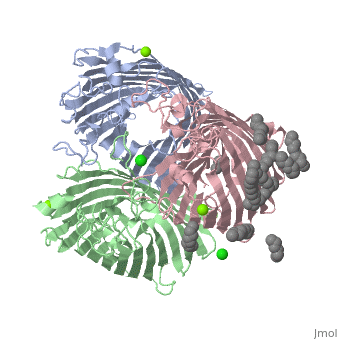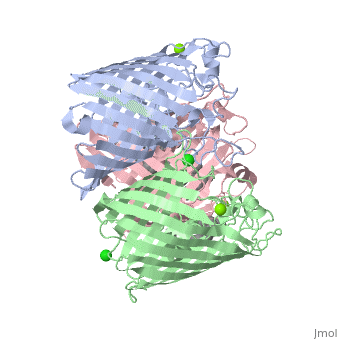
OmpF:4GCP
Certain porins such as 4gcp[6] contain ligands, which operate as it's eyelet. The ligand for 4gcp is an antibody, ampicillin, which is zwitterionic.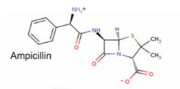
PhoE:1PHO
Unlike the 4gcp porin, some porins contain no ligands to operate as it's eyelet. An example of a porin without a ligand would be the 1pho[8] porin. Instead of having a ligand as it's eyelet, 1pho has a loop inside, which operates as it's constriction zone or eyelet. This loop is slightly anion selective and creates ion selectivity for molecules to pass through the porin. Other features of the 1pho porin is that they are found in E. coli, they are transports phosphate compounds, and they contain only one chain (A). The rest of the features of the 1pho porin are the same as the general structure of porins described earlier.
References
- ↑ http://www.proteopedia.org/wiki/index.php/Osmoporin_OmpC_%28E._coli%29 access 12/9/15
- ↑ “Porin Channel Protein in Escherichia coli Outer Membrane.” Rs.Noda. Web. 4 Nov. 2015. http://www.rs.noda.tus.ac.jp/~biost/OPFU/yama/public_html/study/folding/md/porin/porin.htm
- ↑ “Biophysical Methods Lecture 4: Membrane Proteins – Porins.” Chembio. Department of Chemistry and Biochemistry University of Guelph. Nov. 2013. Web. 4 Nov. 2015. http://www.chembio.uoguelph.ca/educmat/phy456/456lec04.htm
- ↑ “What are the Different Modes of Biotransportation of Drugs – How Drugs Move Across the Cell Membrane?” HubPages. 28 Sept. 2015. Web. 4 Nov. 2015. http://hubpages.com/education/Different-modes-of-biotransport-of-drugs
- ↑ Casiday, Rachel. Frey, Regina. “Transport Across Membranes: Energetics and Pumps/Channels.” Academy of Gifted Learners. Department of Chemistry, Washington University. Web. 4 Nov. 2015. https://academyofgiftedlearners.files.wordpress.com/2014/11/chapter13_f141.pdf
- ↑ Ziervogel, B.K., Roux, B. “4GCP.” RCSB PDB Protein Data Bank. 6 Feb. 2013. Web. 28 Nov. 2015. http://www.rcsb.org/pdb/explore.do?structureId=4GCP
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3545085/figure/F1/ access 12/9/15
- ↑ Schirmer, T., Cowan, S.W., Jansonius, J.N. “1PHO.” RCSB PDB Protein Data Bank. 13 Jul. 2011. Web. 28 Nov. 2015. http://www.rcsb.org/pdb/explore/explore.do?structureId=1pho