Sandbox viralpackagingmotors
From Proteopedia
This page is setup for Ojuewa to build her senior project for OU CHEM 4923
Contents |
Viral Packaging
|
One of the most important stages in the life cycle of all viruses is the encapsidation (packaging) of the viral genome. Many viruses package their genome into preformed capsids using packaging motors powered by the hydrolysis of Adenosine Tris-phosphate (ATP). The hexameric ATPase P4 of dsRNA bacteriophage phi 12, located at the vertices of the icosahedral capsid, is an example of a packaging motor. [1] The P4 protein is a 35-kDa ssRNA packaging ATPase from dsRNA bacteriophages belonging to the Cystoviridae family. ( phi 6 - phi 12). These viruses use a packaging motor such as the RNA packaging motor that is powered by the hydrolysis of ATP to condense the nucleic acid into a confined space. [2].
Structure
The RNA packaging motor is a simple machinery that consists of a portal protein (P4) hexamer. The crystal structure of the hexameric P4 protein is made of three sub-units, one-half of the hexameric molecule (hexamers encircle the crystallographic 2-fold axes). Residues 196-206, 300-306, and the C-terminal segment 322-331are disordered in all subunits. [1]
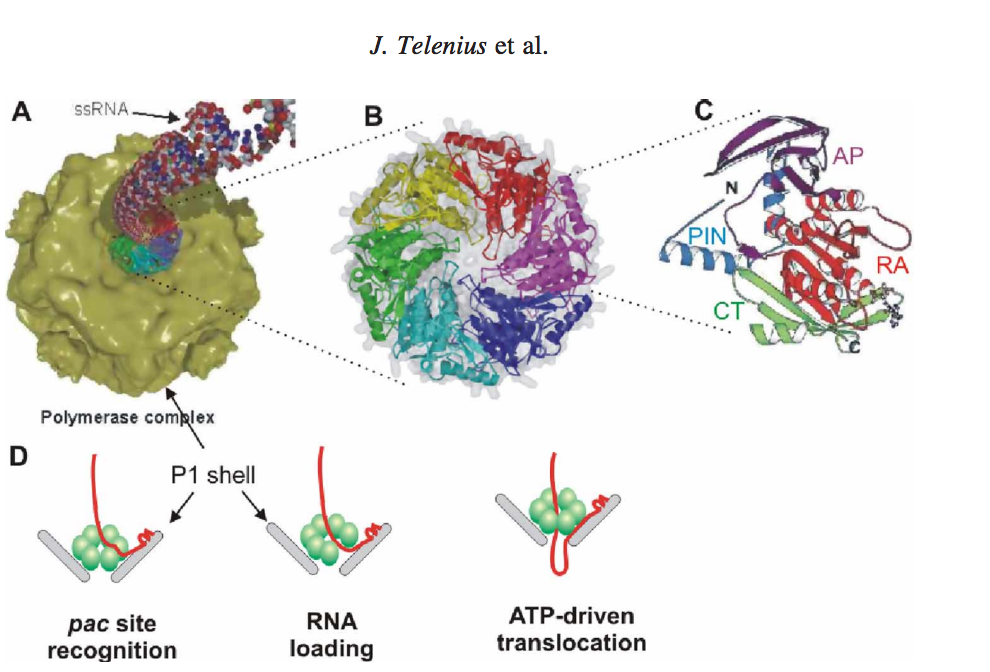 Figure 1.[3]
Figure 1.[3]
Figure 1 shows the mechanism and package of ssRNA precursors into preformed procapsids (also called polymerase complexes).
The mechanism are described as follows: (A) Packaging of ssRNA via P4 hexamer into the icosahedral procapsid (B) P4 hexamer structure (C) Subunit structure: RecA-like catalytic core is shown in red (RA), N-terminal safety pin in cyan (PIN), apical domain in magenta (AP), and C-terminal platform in green (CT). The safety pin stabilizes interacts with a neighboring subunit and stabilizes the hexamer. (D) A scheme of the three step packaging mechanism for a ssRNA precursor. [3].
The P4 viral packaging motor is a sequential molecular machine that must complete several tasks in a sequential order. After successful packaging, the three ssRNAs are converted into dsRNA inside the viral procapsid by a virus-associated RNA-dependent RNA polymerase (protein P2). For each segment, the packaging process may be divided into three steps (Figure 1(D)) which are (i) recognition of the RNA pac site (a packaging signal) by a specific binding site on the surface of the viral capsid (i.e. major structural protein P1), (ii) loading of the ssRNA into the packaging motor (most likely by a ring-opening mechanism), and (iii) unidirectional, 50–30 translocation of the captured ssRNA into the procapsid at the expense of ATP hydrolysis. The dome-shaped P4 hexamer has an overall structure and subunit fold similar to those of hexameric helicases (Figure 1(B)–(C)). The subunit structure has three domains: N-terminal apical dome, middle RecA-like ATPase core, and C-terminal platform. Remarkably, the same basic fold was also found for a packaging ATPase domain from dsDNA bacteriophage T4. [3] [4] [1]. Thus, based on structure alone, it is possible to draw a conclusion that all these motor proteins may share certain mechanistic features. In recent years, the P4 protein has become one of the best characterized hexameric helicases and, given the wealth of structural and biophysical information available, it constitutes a good model system for studying the mechano-chemistry of nucleic acid translocation. [3] [2].
Function
The RNA packaging motor is a straightforward machinery that consists of a portal protein (P4) hexamer. The mechanism of a molecular motor has both the chemical and mechanical reaction coordinates. The energy exchange between the two coordinates (i.e. mechano-chemical coupling) is described. The rules behind the coordinated progress and energy exchange between the two coordinates (i.e. mechano-chemical coupling) is also explained. The X-ray crystallography showed that the movement of a molecular lever was associated with subtle changes in the nucleotide-binding pocket during ATP hydrolysis (Figure 2(A)). The lever consists of two connected elements, an a-helix 6 and a loop named L2. The end of the helix inserts into the ATP binding site of a neighboring subunit, while the loop L2 protrudes into the central channel of the hexamer and returns back to the active site on the same subunit. The L2 loop contains a lysine (Lys241 in f12 P4, Figure 2) which is conserved among most cystoviral P4s. The positively charged side chain interacts with RNA. The position of the L2 loop-a-helix 6 lever, inside the central channel, depends on the occupancy of the ATP binding site. In the presence of AMPcPP, an ATP analogue is in an ‘up’ position while in the ADP-bound state it is in a ‘down’ position (Figure 2(A)). The position of the lever (‘up’ or ‘down’) correlates with the conformation of the P-loop (H1 motif also known as Walker A), which is seen as either retracted from or inserted in the active site (Figure 2(B)). In addition, the ‘down’ position of the side chain of Arg279 (structurally equivalent to ‘arginine finger’ in GAPs and F1-ATPase) is inserted into the neighbouring active site, while it is retracted in the ‘up position’ (Figure 2(E)). In the apo hexamer (i.e. in the absence of bound nucleotide tri- and di-phosphates), the lever can assume both ‘up’ and ‘down’ positions suggesting that the nucleotide occupancy controls this movement. The lever movement shows that it is essential for ATP hydrolysis. Hence, the lever motion could be considered a good approximation for the mechanical reaction coordinate and is tightly coupled to ATP hydrolysis. [3] [1].
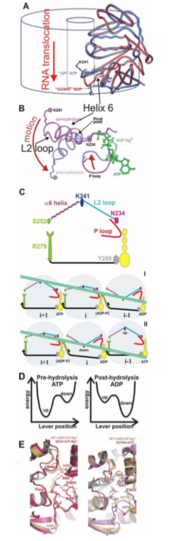
Energetics
A packaging NTpase hexamer (P4) provides energy for RNA translocation. A molecular motor converts chemical energy into mechanical work which is shown by directional motion. How this conversion by molecular motors is achieved is debatable. In essence, the question is how the progress of the chemical reaction (e.g. ATP binding or hydrolysis in the active site of the ATPase) drives or is coupled to the motion along the mechanical coordinate (e.g. RNA translocation through the hexamer) [3]. It is important to know that φ12 p4 (Figure 3) appears to have two arginine fingers residue Arg 272 and Arg 279 located on either side of the a β hairpin. Insertion of this "arginine finger" into the catalytic site is believed to stabilize the transition state, thus facilitating ATP-hydrolysis. These arginines are connected via strand β15 to Try 288; which forms part of an adjacent nucleotide binding site providing a mechanism to transit conformational changes between neighboring active sites upon nucleotide binding and hydrolysis [3] [1].
Structural highlights
The structurally conserved Rec-A like core is made up of five sequence motifs (H1, H1a, H2, H3 and H4) that are common to all superfamily 4 (SF4) helicases (Figure 1(C)) [3]. The sensor motifs that are supposed to detect the presence or absence of γ-phosphate of ATP and relay the information to other sites within the molecule, through conformational changes, were first identified as RecA. The active sites of RecA like ATPases usually contain a second motif contributed from an adjacent monomer, commonly an Arginine residue(Figure 3), containing the γ-phosphate of ATP. [3] [4].
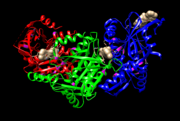
Packaging the genome into an empty procapsid is central to the assembly of many complex virus. Many of these motor proteins from ring-shaped oligomers and are part of an enormous superfamily of molecular motors (AAA+ proteins)[3].
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004 Sep 17;118(6):743-55. PMID:15369673 doi:10.1016/j.cell.2004.09.007
- ↑ 2.0 2.1 Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647-81. doi: 10.1146/annurev.genet.42.110807.091545. PMID:18687036 doi:http://dx.doi.org/10.1146/annurev.genet.42.110807.091545
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 doi: https://dx.doi.org/10.1080/17486700802168502
