User:Alexis Neyman/Sandbox 1
From Proteopedia
Contents |
Biological Structure of SRp20
Introduction
Alternative RNA splicing is a significant post-transcriptional process that allows diversity of gene expression. Initially, a gene is transcribed into pre-messenger RNA (pre-mRNA). The pre-mRNA contains introns, which are sequences that are not translated into protein, and exons, which code for proteins. In alternative RNA splicing, exons are either removed or retained in the mRNA in different combinations, creating diverse arrangements of mRNAs from one pre-mRNA. This process is carried out with splicing factors which are proteins that remove introns and exons via spliceosomes [1]. The sequence-specific RNA binding protein SRp20 (gene name SRSF3) is a splicing factor and one of the smallest members of the serine and arginine-rich (SR) protein family that plays a significant role in alternative splicing of exons [1] [2]. It is a highly conserved Homo Sapien protein[3].
Function
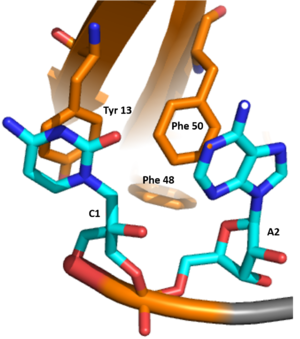
SRp20 is a splicing factor involved in regulation of many genes through alternative splicing of exons by associating with cis-elements of RNA[1][4]. It contains an auto-regulatory activity in which it can alternatively splice its own mRNA by including exon 4 thus reducing the length of its protein[5][6][7]. It has been speculated that SRp20 has been linked to termination of transcription by either activating enzymes responsible for degrading the RNA sequence downstream from the cleavage site or promoting the removal of RNA polymerase from the DNA[8]. SRp20 might play a role in export of mature mRNAs by promoting the recruitment of TAP, which is an export factor for mRNA export out of the nucleus[5]. It has been found that SRp20 and PCBP2, which is a protein that binds to internal ribosome entry site (IRES) RNA sequences in picornavirus, interact with each other to initiate viral translation[9]. Thus, these findings indicate SRp20 plays a role in protein translation. It is also suggested that SRp20 allows 3' terminal exon to be recognized by polyadenylation factors[10].
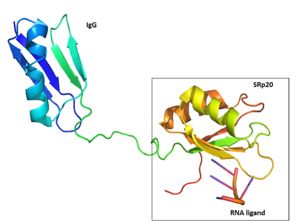
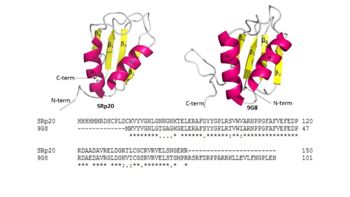
Structure
| |||||||||||
Cancer
There have been findings that support the role of SRp20 in cellular proliferation/maturation. It was discovered that there was an over expression of SRp20 in breast cancer tissues. When SRp20 was reduced in cancer cells via siRNA, targets SRp20 mRNA, there was reduction in cell proliferation and increase in cellular apoptosis. For example, it was speculated that SRp20 might be involved in alternative splicing of FoxM1, a transcription factor involved in cellular proliferation, by either the inclusion or exclusion of exon 9 in FoxM1 transcript. If exon 9 was excluded from the FoxM1 mRNA via SRp20, then there was an increase in FoxM1 expression, cellular proliferation, and reduction in cell apoptosis[3]. Apoptosis is a necessary function to maintain homeostasis, and an imbalance in the regulation in apoptosis can lead to uncontrolled cell proliferation and tumor development. Due to the alternative splicing functionality of SRp20, it effects many other genes involved in cancer such as CD44 gene, PK-M gene, TAU gene, TP53 gene, and involved in WnT signaling pathway[1]. Although it has been understood that SRp20 plays a crucial role in cancer cells, the mechanism by which SRp20 affects these genes, and how its structure contributes to the development of oncogenic genes, is still unclear[3][15].
Relevance and Conclusions
Understanding and recognizing the mechanisms that SRp20 is involved in can help find treatment and management of cancer patients. The use of SR proteins (such as SRp20) may in the future be used for targeted therapy. Because there is no known structure for the C-term domain, due to an inability to obtain a structural image for it, most of the focus has been on the RRM domain. Little is understood about how the SR domain might recognize structures or other proteins.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Corbo C, Orru S, Salvatore F. SRp20: an overview of its role in human diseases. Biochem Biophys Res Commun. 2013 Jun 21;436(1):1-5. doi:, 10.1016/j.bbrc.2013.05.027. Epub 2013 May 16. PMID:23685143 doi:http://dx.doi.org/10.1016/j.bbrc.2013.05.027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006 Nov 1;25(21):5126-37. Epub 2006 Oct 12. PMID:17036044
- ↑ 3.0 3.1 3.2 Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010 Dec 15;6(7):806-26. PMID:21179588
- ↑ Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009 Jan;83(1):167-80. doi: 10.1128/JVI.01719-08. Epub 2008 Oct 22. PMID:18945760 doi:http://dx.doi.org/10.1128/JVI.01719-08
- ↑ 5.0 5.1 Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997 Jun;17(6):3116-24. PMID:9154810
- ↑ Jumaa H, Nielsen PJ. Regulation of SRp20 exon 4 splicing. Biochim Biophys Acta. 2000 Nov 15;1494(1-2):137-43. PMID:11072076
- ↑ Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997 Aug 15;16(16):5077-85. doi: 10.1093/emboj/16.16.5077. PMID:9305649 doi:http://dx.doi.org/10.1093/emboj/16.16.5077
- ↑ Cui M, Allen MA, Larsen A, Macmorris M, Han M, Blumenthal T. Genes involved in pre-mRNA 3'-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc Natl Acad Sci U S A. 2008 Oct 28;105(43):16665-70. doi:, 10.1073/pnas.0807104105. Epub 2008 Oct 22. PMID:18946043 doi:http://dx.doi.org/10.1073/pnas.0807104105
- ↑ Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007 Jan 24;26(2):459-67. doi: 10.1038/sj.emboj.7601494. Epub 2006 Dec, 21. PMID:17183366 doi:http://dx.doi.org/10.1038/sj.emboj.7601494
- ↑ Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998 Sep;18(9):4977-85. PMID:9710581
- ↑ 11.0 11.1 11.2 Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008 Jun;18(3):290-8. doi: 10.1016/j.sbi.2008.04.002. PMID:18515081 doi:http://dx.doi.org/10.1016/j.sbi.2008.04.002
- ↑ Mackness BC, Tran MT, McClain SP, Matthews CR, Zitzewitz JA. Folding of the RNA recognition motif (RRM) domains of the amyotrophic lateral sclerosis (ALS)-linked protein TDP-43 reveals an intermediate state. J Biol Chem. 2014 Mar 21;289(12):8264-76. doi: 10.1074/jbc.M113.542779. Epub 2014, Feb 4. PMID:24497641 doi:http://dx.doi.org/10.1074/jbc.M113.542779
- ↑ Netter C, Weber G, Benecke H, Wahl MC. Functional stabilization of an RNA recognition motif by a noncanonical N-terminal expansion. RNA. 2009 Jul;15(7):1305-13. Epub 2009 May 15. PMID:19447915 doi:10.1261/rna.1359909
- ↑ 14.0 14.1 Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999 Mar;5(3):468-83. PMID:10094314
- ↑ Jang HN, Lee M, Loh TJ, Choi SW, Oh HK, Moon H, Cho S, Hong SE, Kim DH, Sheng Z, Green MR, Park D, Zheng X, Shen H. Exon 9 skipping of apoptotic caspase-2 pre-mRNA is promoted by SRSF3 through interaction with exon 8. Biochim Biophys Acta. 2014 Jan;1839(1):25-32. doi: 10.1016/j.bbagrm.2013.11.006. , Epub 2013 Dec 7. PMID:24321384 doi:http://dx.doi.org/10.1016/j.bbagrm.2013.11.006