User:Satyan Sharma/Sandbox 1
From Proteopedia
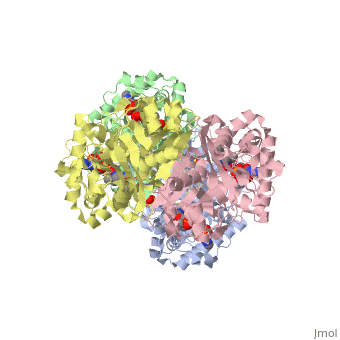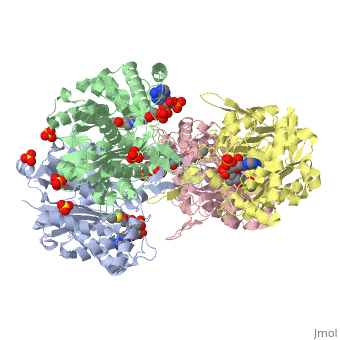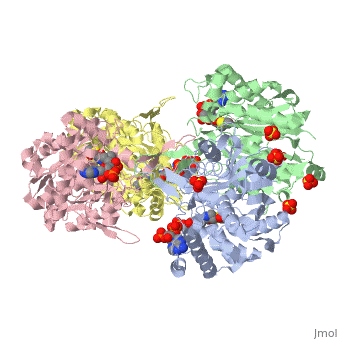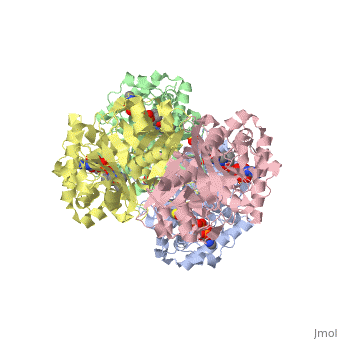
3D structure (1DM3) of the bacterial Zoogloea ramigera biosynthetic thiolase
Contents |
Introduction
Thiolase is best known as a key enzyme in the fatty acid degradation pathway, also known as the β-oxidation pathway. However, in the human metabolism at least six thiolases (Table 1) have been characterized. Each of them important in different pathways, and in different cell organelles. Some thiolases are dimers, some thiolases are tetramers, being dimers of dimers. Thiolases are involved, either in degradative pathways (E.C. 2.3.1.16), or in biosynthetic pathways (E.C. 2.3.1.9). This division is somewhat arbitrary as all thiolases are sequence related to each other and catalyse the reaction in both directions.
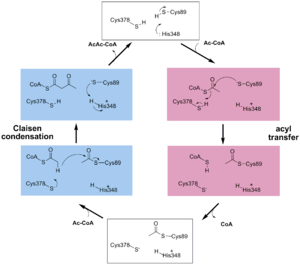
The above scheme shows the degradative reaction catalysed by thiolase. In this the acetoacetyl-CoA is degraded with CoA as cosubstrate, by which two molecules of acetyl-CoA are formed. Each thiolase catalyzes this reaction in both directions, but the
equilibrium is far much favoring the degradative direction
(Keq is 105, favoring the degradation). The
reaction is catalyzed by the enzyme as a two step reaction. In the first step a fully conserved
active site cysteine, Cys89 is acetylated, liberating acetyl-CoA. In
the second step the acetyl-moiety is transferred to CoA.
The best studied thiolase is the bacterial Zoogloea ramigera thiolase, which is a biosynthetic thiolase and which catalyzes the formation of acetoacetyl-CoA from two molecules of acetyl-CoA. The enzymological properties of Zoogloea ramigera thiolase have been extensively characterized by C.T Walsh and coworkers[1] and more recently its structural enzymological properties have been characterized also.[2][3] The residue numbering on this page refers to the Zoogloea ramigera thiolase sequence, and the structural information concerns the 1DM3 structure.
| Intracellular localization | Mode of assembly | Function | Comment: thiolase protein code (gene sequence code) |
|---|---|---|---|
| cytosol | α4 | synthesis | CT (THIC) |
| mitochondria | α4 | synthesis and degradation | T2 (THIL) Many point mutation variants related to metabolic disorders have been characterized.[4] |
| α4 | degradation | T1 (THIM) | |
| α4β4 | degradation | trifunctional enzyme, TFE (ECHB) | |
| peroxisome | α2 | degradation | A/B (THIK) |
| α2 | degradation | SCP2 (NLTP) |
The reaction mechanism
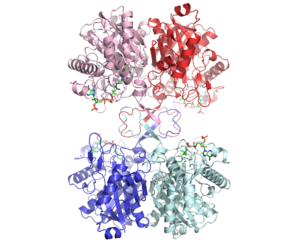
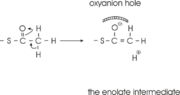
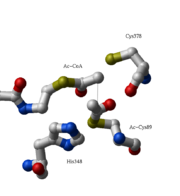
The complete catalytic cycle of the Zoogloea ramigera thiolase is shown in Figure 1. This thiolase is a tetramer, indeed a dimer of dimers, as shown in Figure 2. The active site is deeply buried. It is constructed by , which provide the catalytic nucleophile (Cys89), the base (Cys378), as well as Asn316 and His348. Asn316 and His348 are key residues of the active site geometry: a Asn316-Wat82 dyad, together with His348 make , stabilizing the CoA-thioester enolate intermediate. The hydrogen bond donors of oxyanion hole 1 are Wat82 and NE2 (His348). This enolate is formed from acetyl-CoA after proton abstraction by the catalytic base, Cys378 (Figure 3).
|
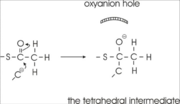
His348 has a dual role, as it also activates Cys89 for nucleophilic attack. A further key element of the active site geometry is , which stabilizes the tetrahedral intermediate thioester oxyanion. This oxyanion hole is formed when the C2-atom of acetyl-CoA reacts with the carbonyl-atom of the acetyl-cysteine thioester (Figure 4). The hydrogen bond donors of oxyanion hole 2 are N (Cys89) and N (Gly380).
The importance of the Asn316-Wat82 diad and His348 for the oxyanion hole 1 stabilization of the transition state in the Claisen condensation reaction has been confirmed by recent studies by Merilainen et. al.[3]
These key elements of the thiolase reaction mechanism have been captured in the 1DM3 crystal structure,[5] which is a complex of acetyl-CoA complexed with the acetylated enzyme (Figure 5). This crystal form is grown at pH5. At this pH the catalytic base, Cys378, is predicted to be always protonated, independent of the ligand bound in the active site. Therefore it is unable to abstract the proton from the methyl group of acetyl-CoA, and therefore the reaction intermediate as shown in Figure 5 has been captured.
There are several structures known of the Zoogloea ramigera thiolase complexed with the intermediates of the reaction cycle.[2] It is interesting to point out that no gross structural changes of loops or side chains have been detected. For example, the only structural change with the apo structure (1DLU) is the small movement of the Cys89 side chain towards the His348 side chain, by which it is assumed that Cys89 is deprotonated and therefore activated.
Diseases
The best characterized thiolase deficiencies concern mutations in the T2 thiolase gene. T2 is important in the synthesis of ketone bodies (in the mitochondria of the liver) as well as for the breakdown of these ketone bodies, for example in the mitochondria of the brain and heart. The T2 deficiency affects the breakdown of these ketone bodies as is detected by accumulation of the oxidized ketone bodies in the blood stream, causing ketoacidosis. This suggests that some other thiolase is able to replace the synthetic, mitochondrial T2 thiolase function, but not the degradation function. The T2 deficiencies have been studied much by Dr. Fukao (Gifu University, Gifu, Japan).[4]
Evolution
Thiolase turns out to be the founding member of a large superfamily of enzymes, referred to as the thiolase superfamily.[6] Key members of this superfamily are the KAS-enzymes. The KAS-enzyme is the first enzyme in the fatty acid synthesis pathway. It has the same fold as thiolase, and also the nucleophilic cysteine is conserved. Also conserved in the KAS enzymes are the two residues which built oxyanion hole 1 in thiolase. It has been noted that in the superfamily the Asn316 and His348 either occur as a NH-pair (like in thiolase), or as an HH-pair, or as an HN-pair, the NN-pair is never observed. Each of the superfamily members also have the fully conserved cysteine corresponding to Cys89. Therefore the members of the superfamily can be divided into three categories, being CNH, CHH, and CHN.[7] In the bacterial thiolase, extensive mutagenesis followed up by structural enzymology[3] has confirmed that the CNH-triad of Zoogloea ramigera is indeed important for the function of oxyanion hole 1.
References
- ↑ Williams SF, Palmer MA, Peoples OP, Walsh CT, Sinskey AJ, Masamune S. Biosynthetic thiolase from Zoogloea ramigera. Mutagenesis of the putative active-site base Cys-378 to Ser-378 changes the partitioning of the acetyl S-enzyme intermediate. J Biol Chem. 1992 Aug 15;267(23):16041-3. PMID:1353760
- ↑ 2.0 2.1 Kursula P, Ojala J, Lambeir AM, Wierenga RK. The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes. Biochemistry. 2002 Dec 31;41(52):15543-56. PMID:12501183
- ↑ 3.0 3.1 3.2 Merilainen G, Poikela VM, Kursula P, Wierenga RK. The thiolase reaction mechanism: the importance of Asn316 and His348 for stabilizing the enolate intermediate of the Claisen condensation. Biochemistry. 2009 Oct 20. PMID:19842716 doi:10.1021/bi901069h
- ↑ 4.0 4.1 Fukao T, Nguyen HT, Nguyen NT, Vu DC, Can NT, Pham AT, Nguyen KN, Kobayashi H, Hasegawa Y, Bui TP, Niezen-Koning KE, Wanders RJ, de Koning T, Nguyen LT, Yamaguchi S, Kondo N. A common mutation, R208X, identified in Vietnamese patients with mitochondrial acetoacetyl-CoA thiolase (T2) deficiency. Mol Genet Metab. 2010 May;100(1):37-41. Epub 2010 Jan 21. PMID:20156697 doi:10.1016/j.ymgme.2010.01.007
- ↑ Modis Y, Wierenga RK. Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase. J Mol Biol. 2000 Apr 14;297(5):1171-82. PMID:10764581 doi:10.1006/jmbi.2000.3638
- ↑ Haapalainen AM, Merilainen G, Wierenga RK. The thiolase superfamily: condensing enzymes with diverse reaction specificities. Trends Biochem Sci. 2006 Jan;31(1):64-71. Epub 2005 Dec 13. PMID:16356722 doi:10.1016/j.tibs.2005.11.011
- ↑ Jiang C, Kim SY, Suh DY. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol Phylogenet Evol. 2008 Dec;49(3):691-701. Epub 2008 Sep 12. PMID:18824113 doi:10.1016/j.ympev.2008.09.002