Cassady sandbox1
From Proteopedia
| |||||||||
| 1pfk, resolution 2.40Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Activity: | 6-phosphofructokinase, with EC number 2.7.1.11 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Phosphofructokinase (PFK) (PDB id 4pfk) is an approximately 300 residue enzyme that catalyzes the phosphorylation of Fructose-6-phosphate (F6P) to Fructose-1,6-bisphosphate (F1,6P) in the third reaction of glycolysis. This enzyme, with four subunits, catalyzes the most highly regulated reaction of glycolysis. Phosphofructokinase is not only the enzyme's name, but also, the fold, superfamily, and family classification name.
Contents |
Role in Glycolysis
Glycolysis is the process of breaking down glucose to make pyruvic acid, which is used in anaerobic respiration or as one of the starting reactants in the citric acid cycle. The process releases some energy but more importantly paves the way for vast amounts of energy to be made through the citric acid cycle. After glucose has been phosphorylated and isomerized to Fructose-6-phospate, PFK begins its work. It phosphorylates the hydroxy group at the number one carbon, which was impossible in glucose. This second phosphorylation by PFK is important because it sets up the six carbon compound to later become high energy. High energy compounds help to drive the endergonic processes of glycolysis through their own exergonic breakdown.[1]. It is important then that PFK makes a bisphosphate compound, because eventually that molecule will be cut in half. Thus, after the action of PFK, the six-carbon compound can be broken into two high-energy three-carbon compounds, which are both ready to move onto the next steps of catabolism. Also, phosphorylating the F6P allows energy to be captured rather than lost as heat.
Structure
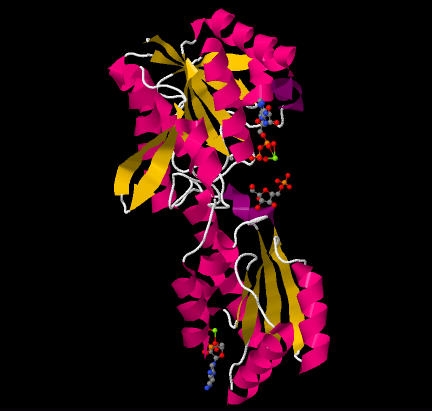
PFK is a that acts as a dimer of dimers, similar to hemoglobin.[2]. One half of each dimer is involved in the binding of ATP, while the other is involved with substrate binding and also contains an allosteric site. [3]. One subunit with ATP and F6P bound can be seen below. PFK's puts it in an alpha and beta class. Each unit is comprised of two domains that sandwich parallel beta sheets in between alpha helices. The outer most beta sheets of the larger domain, however, are anti-parallel. This is best seen in the .
Mechanism
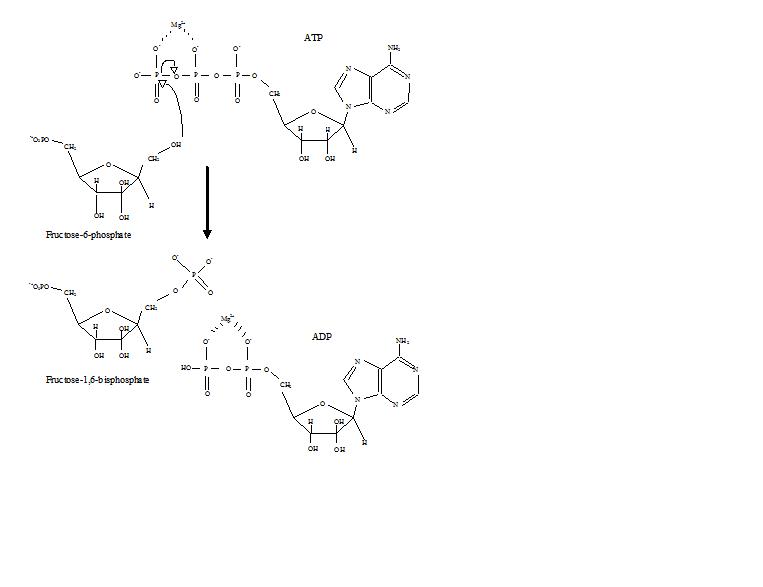
Phosphofructokinase binds both Mg2+-ATP and fructose-6-phosphate (F6P) to make fructose-1,6-bisphosphate and Mg2+-ADP. Although the image with both of these products has not been determined, bound to the enzyme has been. There are three ligand binding sites per subunit. Two make up the active site, which binds F6P and ATP, while the third is an allosteric binding site.[4] Some proposed residues involved at the active site include .[5] PFK exists in two conformational states, both and which are in equilibrium. ATP binds the active and allosteric sites in both conformations.
Regulation
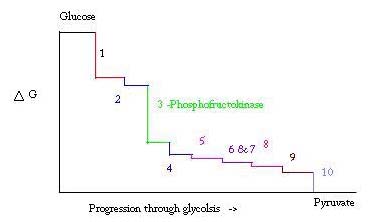
Glycolysis is an essential metabolic process for survival. Therefore, in its activation and suppression it must be highly regulated. Three points in the process of glycolysis occur with a large negative free energy and are therefore, irreversible. These three points are hexokinase, phosphofructokinase, and pyruvate kinase. These three reactions are candidates to be the major points of regulation because of their high negative free energies. Of the three, PFK is considered the major regulatory point for glycolysis (#3 in the picture below) in muscle, with a ΔG= -25.9 kJ/mol, because it is a committed step. Once PFK converts F6P to F1,6P, the reaction will not be easily reversed because of the high amount of energy that must be overcome to go backward. [6]. This energy barrier makes sense seeing as pyruvate kinase catalyzes the final reaction (#10) and hexokinase (#1) is not involved in glycolysis at all when the process is begun from glycogen.[7]
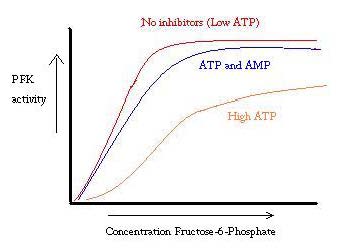
PFK is regulated by ATP, AMP, and ADP. While ATP binds at the active site equally well in both R and T states, it preferentially binds the allosteric site of the T state [8] This preferential binding causes a shift from equilibrium of the two states, to a greater amount of T state [9], which decreases the affinity for F6P. Allosteric activator also binds to the allosteric site to increase the ratio of R state phosphofructokinase. As can be seen from the graph below, the plots for the activity of PFK are sigmoidal. This further demonstrates the cooperative nature of the enzyme. The initial binding of substrate to the enzyme is difficult, but once it is bound and forces the change in state from T -> R, the other substrates bind much more easily. The graph also shows that adding ATP moves the plot right (ie decreases affinity for F6P), while adding AMP moves it to the left.
The system of regulation matches well with the function of PFK. When PFK is active, ATP is being produced down stream from it as further products are broken down more completely. Thus, when ATP levels are low and more needs to be made, the activity of PFK will be increased, because ADP will be in high concentration. The opposite holds true as well, because high ATP concentration inhibits protein activity. And yet, this explanation cannot completely account for the regulation of PFK, because the levels of ATP do not vary greatly enough between active and resting muscles. Another means of allosteric regulation must be found.[10]
PFK's Km for ATP is .020mM and .032mM.[11]
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: structure and control. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):53-62. PMID:6115424
- ↑ Shirakihara Y, Evans PR. Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J Mol Biol. 1988 Dec 20;204(4):973-94. PMID:2975709
- ↑ Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: structure and control. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):53-62. PMID:6115424
- ↑ http://www.nature.com/nature/journal/v327/n6121/abs/327437a0.html
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ PubMed:2136935
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Campos G, Guixe V, Babul J. Kinetic mechanism of phosphofructokinase-2 from Escherichia coli. A mutant enzyme with a different mechanism. J Biol Chem. 1984 May 25;259(10):6147-52. PMID:6233271