Resolution
From Proteopedia
|
Snapshots from the movie (see below) by James Holton. See Limitations of the Movie below. |
Resolution, in structure determinations, is the distance corresponding to the smallest observable feature: if two objects are closer than this distance, they appear as one combined blob rather than two separate objects.
|
Details at [2]. | |
|
Details at [3]. |
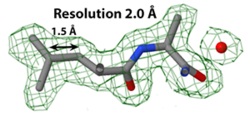
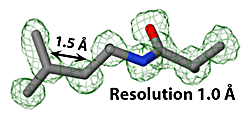
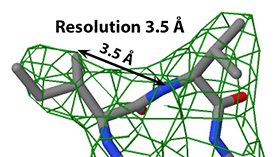
Structure determination by X-ray crystallography or cryo-electron microscopy produces an electron density map (shown in green). The atomic model, shown as sticks, is then built, guided by the electron density map.
In X-ray crystallography, resolution is the smallest distance between crystal lattice planes that is resolved in the diffraction pattern. High numeric values of resolution, such as 4 Å, mean poor resolution, while low numeric values, such as 1.5 Å, mean good resolution.
2.05 Å is the median resolution for X-ray crystallographic results in the Protein Data Bank (135,762 on May 19, 2019). For comparison, the van der Waals diameter of a carbon atom is 3.4-3.7 Å[4], and the length of a covalent carbon-carbon bond is 1.5 Å[4].
If some portions of the macromolecule are less ordered in the crystal than others, these will have a poorer resolution. The "resolution of the crystal" represents the most ordered portions (see Determination of Resolution).
After an electron density map is calculated and refined with a fitted atomic model, an uncertainty of atomic position is calculated for each atom in the model. These single-atom uncertainties are called the B factors or temperature values of the atoms (see Temperature).
Quick Guide
Here is a quick guide for interpreting values of resolution (taken from FirstGlance in Jmol).
- < ~1.7 Å: Most atoms very clearly seen.
- ~1.7 - 2.2 Å: Many atoms clearly seen.
- ~2.2 - 2.8 Å: Shapes of larger sidechains seen.
- ~2.8 - 3.5 Å: Secondary structure & bulky groups seen. Atoms unclear.
- ~3.5 - 4.0 Å: Most chains traceable. Atoms unclear.
- ~4.0 - 6.0 Å: Some chains traceable. Atoms unclear.
- ~6.0 - 8.0 Å: Shapes of domains at best. Atoms unclear.
- > ~8.0 Å: Largest features seen roughly. Atoms unclear.
Bear in mind that the resolution value represents the "best" (most ordered) parts of a crystal, and that other parts may have poorer quality. The temperature value for each atom in a crystallographic model indicates the clarity of its electron density map.
Confusion of high vs. low resolution
High resolution is characterized by being able to distinguish smaller features, so there is an inverse relationship between the quality of a structure and the length value given for the resolution. For example, a 1.0 Å structure resolves finer detail than a 4.0 Å structure, so the 1.0 Å structure is said to have higher resolution than the 4.0 Å structure. For non-experts, it would be less confusing if the terms were fine and rough resolution rather than high and low resolution, but high and low are the established terms in the field.
What Limits Resolution?
In crystals of macromolecules, it is the degree of order in the crystal ("quality of the crystal") that limits the resolution of X-ray crystallography. Resolution is theoretically limited by the wave length of X-rays (on the order of 1 Å), but in practice, the quality of the available crystals determines resolution. More than half of the crystals obtained from various purified proteins are not of "atomic resolution", that is, the disorder it too great to permit determination of molecular structure. Perhaps the greatest challenge facing crystallographers is to obtain a well-ordered crystal that diffracts to "atomic resolution".
The resolution of a diffraction pattern depends on how ordered the crystal is. If it is highly ordered (atoms are in defined positions throughout the crystal and over time), the crystal will diffract to high resolution. The more disorder there is (atoms moving over time, or the content of one unit cell differing from the next), the lower the resolution of the diffraction image. The intensity of diffraction spots drops with increasing disorder. In order to be able to measure weak reflection spots, it is sometimes possible to increase the intensity of the X-rays used in the experiment, increase the exposure time or sensitivity of the detector, or increase the size of the crystals.
The higher the resolution, the more reflections in the data set. The number of reflections increases with the inverse cube of the resolution, so a 1.0 Å data set has eight times the number of reflection spots present in a 2.0 Å data set.
Electron Density Map vs. Resolution
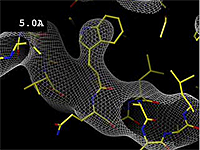
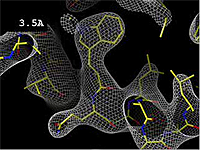
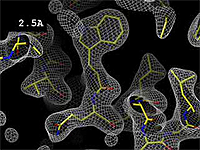
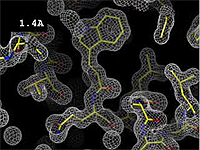
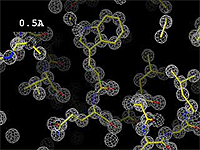
The images at right (top of this page) show how the electron density map[5][6] becomes more accurate and detailed as the resolution value decreases from 5.0 Å to 0.5 Å.
The images at right were taken from a movie[7] in which the atomic model and electron density map rock back and forth while the resolution value (uncertainty) increases from 0.5 to 5.0 Å.
Movie by James Holton at the Advanced Light Source of the Berkeley Laboratory at the University of California[7].)
This movie is not entirely realistic. Limitations are described in the footnote[6].
At 0.5 Å in the movie, every atom[6] of the tryptophan sidechain in the top center of the frame is clearly represented by a sphere of electron density. At 2.5 Å (a bit worse than the median in the PDB), the overall shape and position of the Trp sidechain is still clear, as is the alpha helical conformation of the main chain. However, at 5.0 Å, only an ill-fitting bump is present to signal the bulky Trp sidechain, and the alpha helix becomes a cylinder of electron density, from which the handedness of the helix may not be discernible.
Once refinement of the electron density map is completed, the amount of electron density at the position of each atom determines that atom's B factor or temperature value.
Resolution and structure quality
The higher the resolution of the diffraction data, the more measurements are present to base the model on. Also, an increase in resolution means that a given reflection is measured with less error than in a corresponding lower quality diffraction data set. As a consequence, the model can be built with fewer systematic errors (such as missing or misplaced atoms) and with less average coordinate error. The coordinate error (roughly defined as how different two models would be based on the same crystals, but independent measurement, model building and refinement) correlates with resolution, but is of a different order of magnitude. It is also influenced by completeness of the data (higher is better), the free R-factor of the refinement (lower is better), and the completeness of the model (higher is better). A typical crystallographic model based on 2.0 Å data has a coordinate error of less than 0.2 Å[8].
See also B factors and coordinate error.
Determination of Resolution
The portion of the macromolecule that is best ordered in the crystal is responsible for diffraction spots that are farthest from the axis of the X-ray beam. Resolution is determined from these farthest spots based on their angle θ from the X-ray beam, using the Bragg equation solved for d:
d = λ / (2 sin θ)
where λ is the X-ray wavelength, and d is the smallest distance between crystal lattice planes that scatter discrete spots. The resolution is given by d.
If some portions of the macromolecule are less ordered in the crystal than others, these will have a poorer resolution. The "resolution of the crystal" represents the most ordered portions.
Resolution of a reflection vs. resolution of a diffraction data set
Each diffraction spot (also called "reflection" or "measurement") in a diffraction pattern has a nominal resolution. The higher the diffraction angle of each spot (i.e. the further from the center of the diffraction image where the incoming X-ray beam would hit), the higher is its resolution. In a diffraction experiment, the goal is to collect as many reflections as possible. However, reflections of high resolution are more difficult to measure because the intensity of reflections drops off at higher diffraction angles (and with that, higher resolution). In a diffraction image, you will see high intensity spots near the center, and more and more faded spots as you move away from the center. The overall resolution of a diffraction data set refers to the resolution range of reflections measured. For example, "data were collected from 20.0 Å to 2.3 Å with an overall completeness of 96.5%" means that most reflections in this range were collected, and the data set would be described in brief as "2.3 Å resolution" data set, referring to the high resolution limit of the data collection.
See Also
Within Proteopedia:
- Cryo-EM#Resolution
- Missing Residues and Atoms
- Temperature value
- B factors and coordinate error
- X-ray crystallography
- R value
- Free R
- Hydrogen in macromolecular models
- Believe It or Not
External Resources:
- A Glossary of Terms from Crystallography, NMR, and Homology Modeling
- Resolution (electron density) in Wikipedia.
Notes
- ↑ Slabbed image from Electron Density at 1.0 σ showing Leu425 and Ala426 in 1io1. Temperature factors in those two residues range from 12 to 17, average 15. Separate red atom at right is a water oxygen.
- ↑ Slabbed image from Electron Density Map at 1.0 σ showing Leu5 and Ala4 in 3hyd. Temperature factors in those two residues range from 3 to 12, average 5.
- ↑ Slabbed image showing electron density map at 1.0 &sigma: Ile98 and Val99 in 4gcf. Temperature factors in those two residues range from 32 to 45, average 37.
- ↑ 4.0 4.1 See Atomic radii of the elements (Wikipedia), and values from an Inorganic Chemistry textbook.
- ↑ Electron density maps are the results of X-ray crystallography experiments.
- ↑ 6.0 6.1 6.2 These are "perfect" electron density maps calculated from the atomic model (R factor = 0.0%, perfect phases and amplitudes, contoured at 1 sigma). Electron density maps based on experimental data would fit the true conformation less well. Because these electron density maps were calculated from an atomic model that lacked hydrogen atoms, the electron densities for hydrogen atoms that would appear with experimental data at a resolution of 0.5 Å do not appear.
- ↑ 7.0 7.1 The movie (Image:Resolution holton.mpeg) was created by James Holton at the Advanced Light Source of the Berkeley Laboratory at the University of California. Holton kindly gave explicit permission to use this movie in Proteopedia. The original source was http://ucxray.berkeley.edu/~jamesh/movies.
- ↑ Garib N. Murshudov and Eleanor J. Dodson, Simplified error estimation a la Cruickshank in macromolecular crystallography, CCP4 Newsletter, January, 1997.
Authorship and Acknowledgements
This article was extensively revised June 15, 2014 by User:Eric Martz. The revision was inspired by discussions with Keiichi Namba. Much of the text of this revision was authored by User:Karsten Theis, who put his contributions initially in the discussion page.
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Joel L. Sussman, Wayne Decatur, Eran Hodis, YongLiang Jiang, Jaime Prilusky