Sandbox 150
From Proteopedia
Contents |
Background
Glycolysis is a 10-step process that invests energy in the initial stages only to recover greater amounts of energy in the final steps. Every step in this metabolic pathway is essential to the ultimate production of energy. Every step is catalyzed by one or more enzymes that enhance the rate of the given reaction. Phosphoglycerate mutase is the specific homotetramer enzyme that catalyzes step 8 of glycolysis transfering the phosphate from 3-phosphoglyceric acid to the second carbon to form 2-phosphoglyceric acid, having the Protein Data Bank ID 1qhf[1]. Phosphoglycerate mutase (PGM) is found in organisms from yeast to humans because it plays a significant role in glycolysis, which is a highly conserved process across many taxa. A deficiency of this enzyme causes CNS symptoms, muscle weakness, cramps and fatigue with exercise. [2]
Structure
In terms of the , this protein is classified as an alpha/beta protein. Further, the fold is classified as “phosphoglycerate mutase-like”, having 3 main layers of alpha/beta/alpha. PGM contains a mixed beta sheet of 6 strands, with strand 5 existing as an anti-parallel strand to the rest. The quaternary structure usually is comprised of two identical subunits, thus this enzyme can be classified as a homodimer. The dimers have a relative molecular mass of 56,000-60,000 kDa. [3] One exception includes the PGM enzyme of yeast which is a of mass 110,000 kDa. [3] Though the quaternary structure is the same in terms of the active site, several variations exist, called isozymes, which depend on the tissue in which the enzyme is active. Mm-type, mb-type, and bb-type are isozymes that catalyze glycolysis in smooth muscle, cardiac and skeletal muscle, and the remaining tissues, respectively.[4]
| |||||||||
| 1qhf, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Phosphoglycerate mutase, with EC number 5.4.2.1 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Reaction and Mechanism
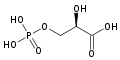
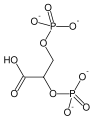
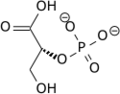
PGM is an integral step in the process of glycolysis. Since this enzyme is a mutase, it will catalyze the transfer of a functional group from one position to another on a given substrate makin this an isomerization reaction. It is responsible for the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG), having 2,3-bisphosphoglycerate as an intermediate. [5] With a Gibbs free energy of about 1.1 KJ/mol, this reaction is nearly energetically neutral. Despite this, it is absolutely necessary in order to generate the proper molecule needed to continue in the glycolytic pathway. The reaction that PGM catalyzes is shown below:
3PG + P-Enzyme → 2,3BPG + Enzyme → 2PG + P-Enzyme 3-phosphoglycerate intermediate 2-phosphoglycerate
It is important to note that the phosphate group that is placed on C2 is not the same phosphate group that was initially on C3. In order to understand how PGM catalyzes this reaction, an explanation of its active site is imperative. The most important residues in this enzyme include with imidazole groups which are in close proximity to carbons 2 and 3 in the substrate. His-8 is phosphorylated during during catalysis, and it is likely that His-179 acts as the proton donor/acceptor [6]. Based on crystallography experiments, the active site where these histidine residues reside lies at the bottom of a deep groove in each subunit. [3] The sites in each subunit, whether the enzyme is a homodimer or homotetramer, are well separated. The active enzyme contains a phosphoryl group attached to His 8. This phosphoryl group is what is transferred to C2 of the substrate, resulting in an intermediate 2,3-bisphosphoglycerate-enzyme complex. Thus there is a in the active monomer. [5] The phosphate group on C3 of the substrate is then transferred back onto His 8, thus regenerating the active form of the enzyme.
In addition to the importance of the two histidine residues in the active site, the amino acids that line the are also functionally important. These residues include H179, H8, E15, S11, T20, R59, and E86.[5] Several positively charged residues line the active site pocket. These residues usually tend to be , which are important for the optimal activity of the enzyme. [3] This structure is logical for its function because the enzyme binds a negatively charged substrate, thus a positively charged groove fosters tight binding with a negative substrate. The third and final important aspect of the active site is the presence of .[3] It is suggested that the carboxyl groups of these amino acid residues act as proton-withdrawing groups as they flank both sides of the substrate.
Kinetics
According to kinetics studies, both cofactor independent and dependent phophoglycerate mutases were found to follow Michaelis-Menten kinetics. Km values were found to be from about 100-200 μM. Other studies reveal how salt concentration and pH influence the kinetics of phosphoglycerate mutase. At low ionic concentrations Km values were around 1 μM, but increased to about 40 μM with the addition of 400 mM KCl. The presence of negatively charged ions in solution could possibly compete with the negatively charged substrate, thus increasing the Km. Additionally, the pH optimum for the enzyme from all species has been found to be 5.9. [3]
Regulation
In terms of regulation, competitive inhibitors resemble the negatively charged substrate and bind to the active site. Such inhibitors include inositol hexakisphosphate and benzene hexacarboxylate. [7] Additionally, the phosphomethyl analogue of 3-phosphoglycerate (2-hydroxy-4-phosphonobutanoate) is a potent inhibitor of phosphoglycerate mutase. [8] These along with many other polyanions, including EDTA, have been reported to act as competitive inhibitors of phosphoglycerate mutase due to their anionic resemblance. As mentioned before, phosphoglycerate mutase has a rather small positive Gibbs free energy. Thus, this reaction proceeds easily in both directions. Overall, this reaction is not the site of major regulation because it is a reversible reaction.
Phosphoglycerate Mutase Deficiency
When phosphoglycerate mutase has a genetic defect, it results in a muscle disease that interferes with the processing of carbohydrates. The onset can occur anywhere from childhood to adulthood. The inheritance pattern is autosomal recessive. [9] Phosphoglycerate mutase deficicncy patients may experience CNS symptoms such as mental retardation and seizures. Certain individuals may experience a purely myopathic syndrome with progressive proximal muscle weakness and incidents of myoglobinuria, exercise intolerance, may become easy fatigued with cramps and urine discoloration. Diagnosing this deficiency can be done with Laboratory tests that demonstrate and increased serum CK level. Or there are diagnostic tests available that test for the absence of the enzyme. Also, muscle pathology of this deficiency shows subsarcolemmal glycogen ± tubular combinations. [10]
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Crowhurst GS, Dalby AR, Isupov MN, Campbell JW, Littlechild JA. Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1822-6. PMID:10531478
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 S., Winn I., Fothergill A. L., Harkins N. R., and Watson C. H. "Structure and Activity of Phosphoglycerate Mutase." Sciences 293.1063 (1981): 121-30. Print.
- ↑ "Phosphoglycerate mutase -." Wikipedia, the free encyclopedia. Web. 27 Feb. 2010. <http://en.wikipedia.org/wiki/Phosphoglycerate_mutase>.
- ↑ 5.0 5.1 5.2 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. Print.
- ↑ Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253
- ↑ Rigden, D. J.; Walter, R. A.; Phillips, S. E. V.; Fothergill-Gilmore, L. A.Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis J. Mol. Biol. 1999, 289, 691– 699
- ↑ McAleese, S.M., Fothergill-Gilmore, L.A.&Dixon, H.B.F. (1985) Biochem. J. 230, 535-542
- ↑ http://www.mda.org/disease/pgam.html
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45