Sandbox Reserved 323
From Proteopedia
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
| |||||||||
| 1k7l, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Related: | 1k74, 1fm6, 1fm9 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
PPAR alpha
Contents |
Introduction
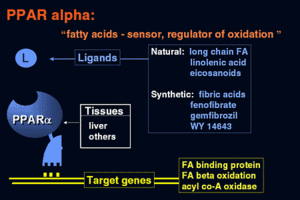
Peroxisome Proliferator-Activated Receptors (PPARs) consist of a superfamily of nuclear hormone receptors that act as transcription factors. PPARs are ligand-activated transcription factors that govern numerous biological processes, including energy metabolism, cell proliferation and inflammation [1]. PPAR-alpha is one of the three PPAR subtypes (alpha, beta and gamma), which can be distinguished by its activating ligand and its specific PPAR-regulated genes to which it provides transcriptional control. PPAR-alpha has an important role in the regulation of nutrient metabolism, including fatty acid oxidation, gluconeogenesis and amino acid metabolism [2]. PPAR-alpha is predominately expressed in tissues with high rates of mitochondrial beta oxidation, such as liver, heart, muscle, kidney and cells of the arterial wall [3]. Various activating molecules of PPAR alpha have consisted of fibrates, fatty acids and eicosanoids, 15-d Ptg (15-Deoxy-Delta Prostaglandin-J2) and oxidized fatty acids. As PPAR-alpha (as well as the other associated isotypes) has an important role in the regulation of energy metabolism, and as its activity can be manipulated by various drugs, there is an increasing interest in the potential connection between PPAR alpha and obesity [1].
Structure
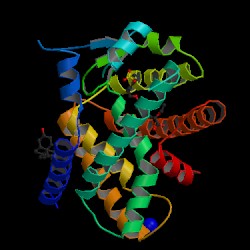
|
The structure shares common characteristics with the other isoforms in this nuclear receptor superfamily, as it displays five distinguishable domains designated the A/B, C, D, E and F domains. The A/B domain, on the N-terminus of PPAR-alpha, contains an activation function region (AF-1), which has a low level of basal transcriptional activity and functions independently of ligand-binding. (C) contains two very highly conserved zinc finger motifs and architectural elements capable of sequence-specific binding to DNA [4]. A (D) connects the DNA binding domain to the ligand-binding domain (E). The ligand-binding domain in the human PPAR alpha protein contains the activation function-2 (AF-2) region composed of two alpha helices flanking one four-sided beta sheet. Following ligand interaction, the AF-2 domain undergoes a conformational change which promotes the hydrogen bonding between , as well as the formation of the “charge clamp” between .[4]
Ligand Interactions
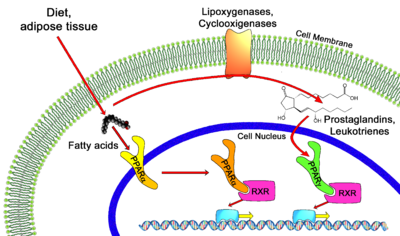
PPAR alpha ligands can consist of either synthetic drugs (exogenous) or biological molecules (endogenous). Most common biological molecules that serve as ligands for human PPAR alpha are fatty acids and fatty acid derivatives [1][2]. Several enzymes such as 8-, 12-, 15-, and 5-lipoxygenases, the cyclooxygenases and cytochrome P450 utilize fatty acids as substrates to produce putative PPAR alpha ligands. Leukotriene B4 is a well-known putative PPAR alpha ligand, as well as 19- and 20- hydroxyeicosatetraeonic acid, generated from cytochrome P450 as a conversion of arachidonic acid. Oleylethanolamide, a naturally occurring lipid, has also been identified as a high affinity PPAR alpha ligand regulating PPAR alpha activity and lipid metabolism [5].
Proliferation of peroxisomes in liver parenchymal cells and upregulated transcriptional activity of fatty acid oxidation system genes are the general hallmarks of exogenous ligand-induced responses, as observed in rat and mouse liver. These include, but not limited to, hypolipidemic drugs such as clofibrate, fenofibrate, bezafibrate, and ciprofibrate [5].
Within the ligand-binding domain (LBD) of PPAR alpha is a large pocket of approximately 1,400 Å, into which a small molecule ligand is bound. The ligand adopts a conformation within the receptor that allows an acidic head group to form hydrogen bonds with Tyr-314 and Tyr-464 on the AF-2 helix [6]. These interactions stabilize the AF-2 helix in a conformation that generates the “charge clamp.” A major determinant of selectivity between the PPAR isotypes alpha and beta is the substitution or Tyr-314 in PPAR alpha for His-323 in PPAR beta. Although all three PPAR subtypes bind to polyunsaturated fatty acids with micromolar affinity, only PPAR alpha has been observed to bind a wide range of saturated fatty acids [6].
Function
The common functionality between the three isoforms consists of binding with a heterodimeric protein, Retinoid X Receptor (RXR) [7][8], to regulate gene expression by recognizing specific DNA elements known as Peroxisome Proliferator Response Elements (PPREs). The heterodimeric transcription factor complex then binds to cognate sequences in promoter regions of target genes involved in the catabolism of fatty acids [8]. Transcriptional activation or repression of the target gene is more complex than simple binding of the PPAR/RXR heterodimer, there is an initial association with transcriptional corepressors and recruitment of activators that occurs in association to ligand activation [2][6][8].
PPAR-alpha is involved in regulating the expression of genes involved in the peroxisomal and mitochondrial Beta oxidation pathways such as Acyl-CoA oxidase, Enol-Co dehydrogenase/hydratase, Malic enzyme and various others [8]. Activated PPAR-alpha contributes to increased breakdown of triglycerides and fatty acids, increased uptake of cellular fatty acids, and reduction in triglyceride and fatty acid synthesis. PPAR alpha displays the greatest amount of activity in the post-absorptive state or fasting state within animals. In the liver, fatty acids are oxidized to acetyl-CoA and/or ketone bodies, in which both processes are strongly stimulated by the expression of PPAR alpha [7]. Since fatty acids act as ligands of PPAR alpha, it is possible that fatty acids liberated from adipose tissue can stimulate their own metabolism by activating PPAR alpha receptors.
All three isoforms of PPAR are becoming more and more of an interest in scientific communities as possible mechanisms of treatment in various diseases and metabolic disorders such as Diabetes Mellitus and obesity. Fibrates, which include fenofibrate, bind with PPAR alpha in high affinity and act to lower plasma triglyceride levels and increase high density lipoprotein levels[9]. Therefore, fibrates may also have an anti-diabetic effect as a consequence of their hypolipidaemic action. PPAR alpha may also have a hand in the prevention of atherosclerosis by decreasing the plasma concentrations of pro-atherosclerosis proteins such as fibrinogen and C-reactive protein [9]. Much of the current research involved with PPARs is directed towards the identification of highly specific agonists and antagonists for treatment of numerous metabolic diseases and conditions.
Additional Resources
- See: Peroxisome Proliferator-Activated Receptors for additional information on PPARs
- See: PPAR-gamma for additional information on another isoform of PPAR
- See: Diabetes Mellitus for additional information on the pathophysiology of Diabetes
References
- ↑ 1.0 1.1 1.2 Kersten S. Peroxisome proliferator activated receptors and obesity. Eur J Pharmacol. 2002 Apr 12;440(2-3):223-34. PMID:12007538
- ↑ 2.0 2.1 2.2 Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999 Jun;103(11):1489-98. PMID:10359558 doi:10.1172/JCI6223
- ↑ Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, Teissier E, Minnich A, Jaye M, Duverger N, Brewer HB, Fruchart JC, Clavey V, Staels B. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med. 2001 Jan;7(1):53-8. PMID:11135616 doi:10.1038/83348
- ↑ 4.0 4.1 Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999 Oct;20(5):649-88. PMID:10529898
- ↑ 5.0 5.1 Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal. 2010 Apr 16;8:e002. PMID:20414453 doi:10.1621/nrs.08002
- ↑ 6.0 6.1 6.2 Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT Jr, McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13919-24. Epub 2001 Nov 6. PMID:11698662 doi:10.1073/pnas.241410198
- ↑ 7.0 7.1 Libby P. Current concepts of the pathogenesis of the acute coronary syndromes. Circulation. 2001 Jul 17;104(3):365-72. PMID:11457759
- ↑ 8.0 8.1 8.2 8.3 Roberts-Thomson SJ. Peroxisome proliferator-activated receptors in tumorigenesis: targets of tumour promotion and treatment. Immunol Cell Biol. 2000 Aug;78(4):436-41. PMID:10947870 doi:10.1046/j.1440-1711.2000.00921.x
- ↑ 9.0 9.1 Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000 May 25;405(6785):421-4. PMID:10839530 doi:10.1038/35013000