Tom sandbox1
From Proteopedia
Contents |
Succinate Dehydrogenase
| |||||||||
| 2wdv, resolution 3.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , , , | ||||||||
| Activity: | Succinate dehydrogenase (ubiquinone), with EC number 1.3.5.1 | ||||||||
| Related: | 2wdq, 2wdr, 1nek, 1nen, 2acz | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The structure of Escherichia coli succinate dehydrogenase (SQR), analogous to the mitochondrial respiratory complex II, has been determined, revealing the electron transport pathway from the electron donor, succinate, to the terminal electron acceptor, ubiquinone. It was found that the SQR redox centers are arranged in a manner that aids the prevention of reactive oxygen species (ROS) formation at the flavin adenine dinucleotide. This is likely to be the main reason SQR is expressed during aerobic respiration rather than the related enzyme fumarate reductase, which produces high levels of ROS. Furthermore, symptoms of genetic disorders associated with mitochondrial SQR mutations may be a result of ROS formation resulting from impaired electron transport in the enzyme.
Architecture of succinate dehydrogenase and reactive oxygen species generation., Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S, Science. 2003 Jan 31;299(5607):700-4. PMID:12560550
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
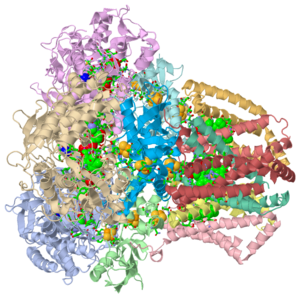
Succinate Dehydrogenase (PDB = 2wdv with an empty ubiquinone binding site; PDB = 1nek with ubiquinone bound), also called succinate-coenzyme Q reductase (SQR) or Complex II, is a tetrameric enzyme. It is found in the cell membrane of some bacteria and the inner mitochondrial membrane of mammalian cells. Classified as an α + β protein, it contains of α helices and antiparallel β sheets. Succinate Dehydrogenase catalyzes the oxidation of succinate to fumarate in the citric acid cycle by simultaneously reducing ubiquinone to ubiquinol via the electron transport chain [1]. This process generates FADH2 after FAD adds on to a Histidine residue in SdhA, one of the Succinate Dehydrogenase subunits. FADH2 goes on to generate massive amounts of ATP. Succinate Dehydrogenase is the only membrane-bound enzyme of the citric acid cycle, so it is positioned to funnel electrons directly into the electron trasport machinery of the mitochondrial membrane[2].Each of these reactants is only a piece of the respiration pathway, the main energy generating pathway for humans and many other species. Succinate Dehydrogenase has been a source of new study after mutation studies have shown it to be the cause of Leigh Syndrome, Familial Paraganglioma, Infantile Leukoencephalopathy, and other cancers [3] [4].
Structure
Hydrophilic Subunits A and B
The quaternary tetramer structure is composed of . The hydrophilic subunits are named SdhA (PDB = 2wdq) and SdhB. SdhA is a flavoprotein, meaning it contains a nucleic acid derivative of riboflavin. SdhA is the longest of the four subunits and is 588 amino acids long. It contains a covalently-bound FAD cofactor and a binding site for succinate as it is the only subunit responsible for the dehydrogenation of succinate to fumarate. SdhB is the other hydrophilic subunit, characterized by the Fe-S bonds, bearing the three iron-sulfur clusters 2Fe-2S, 3Fe-4S, and 4Fe-4S. These iron binded sites can be seen to the right in the Jmol image. SdhB is 238 amino acids long.
Hydrophobic Subunits C and D
The hydrophobic subunits, termed SdhC and SdhD, anchor the protein in the hydrophobic fatty acid portion of the mitochondrial membrane and formally comprise cytochrome b [5] [6]. Each of these subunits is considerably smaller than their hydrophilic partners at 129 amino acids for SdhC and 115 for SdhD. This cytochrome they generate contains six transmembrane α-helices, a heme b group, and a binding site for ubiquinone located in a space bounded by subunits SdhB, SdhC, and SdhD [6] [7]. The cytochrome generated between these subunits is the functional electron transport unit of Succinate Dehydrogenase. The dehydrogenation of succinate works as an electron donor, while the reduction of ubiquinone is the electron acceptor.
Binding sites
Succinate
The binding site for the stereospecific catalyzed dehydrogenation of succinate to fumarate is located entirely on SdhA. stabilize the substrate with hydrogen bonding, while FAD removes the electrons and carries them to the first iron-sulfur cluster, 2Fe-2S, of SdhB [8]. During this transfer, FAD is reduced to FADH2 [2].
Ubiquinone
The binding site for the reduction of ubiquinone to ubiquinol is bordered by subunits B, C, and D. Residues His207 of SdhB, Ser27 and Arg31 of SdhC, and Tyr83 of SdhD stabilize ubiquinone. Residues Pro160, Trp163, Trp164, and Ile209 of SdhB and Ser27 and Ile28 of SdhC provide the necessary hydrophobic environment that stabilizes the ring [7]. In the associated figure from PDB 1nek, the as follows: orange indicates the FAD cofactor, green shows the Fe-S clusters, cyan indicates ubiquinone in its binding site, yellow shows Ca2+ ions, navy blue indicates oxaloacetate, pink is cardiolipin, brown is ephrin, and the heme group is indicated by the lime green structure. The exact function of some of these ligands with regard to succinate dehydrogenase remains unclear. Ephrin, for example is suspected to be involved in certain cell signaling pathways in animal development [9].
| |||||||||
| 1nek, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , , , , , , | ||||||||
| Gene: | SDHA OR B0723 OR Z0877 OR ECS0748 (Escherichia coli), SDHB OR B0724 (Escherichia coli), SDHC OR CYBA OR B0721 OR Z0875 OR ECS0746 (Escherichia coli), SDHD OR B0722 (Escherichia coli) | ||||||||
| Related: | 1nen | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Mechanisms
The Km for the coupled reaction of oxidizing succinate and reducing ubiquinone is approximately 10E(-3)M, while the Vmax is approximately 100 nmol/min/mg of protein [10]
Succinate oxidation
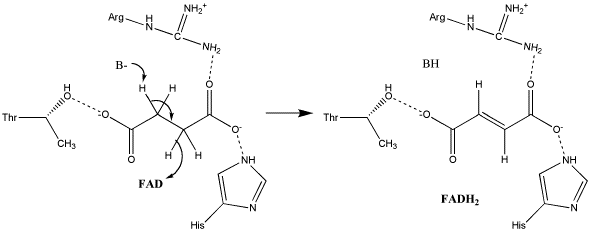
The exact mechanism for the oxidation of succinate to fumarate has not yet been discovered. The initial deprotonation may be performed by FAD, Glu255, Arg286, or His242 of SdhA, and the following elimination may be a concerted E2 or E1cb elimination. In the concerted mechanism, the α-carbon is deprotonated by a base as FAD removes a hydride from the β-carbon; this is shown in image 1 [11].
Image 1: Oxidation of succinate to fumarate through E2 elimination (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:S.D.Oxidation_of_Succinate_E2.gif).
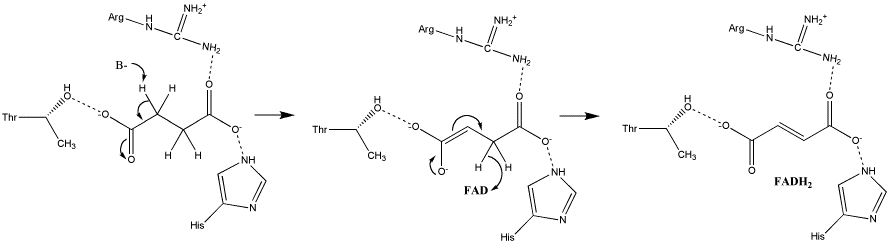
In the proposed E1cb mechanism, the deprotonation leads to the formation of an enolate intermediate; FAD then removes the hydride, as shown in Image 2 [11].
Image 2: Oxidation of succinate to fumarate via E1cb elimination (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:S.D.Oxidation_of_Succinate_E1cb.gif).
Ubiquinone reduction
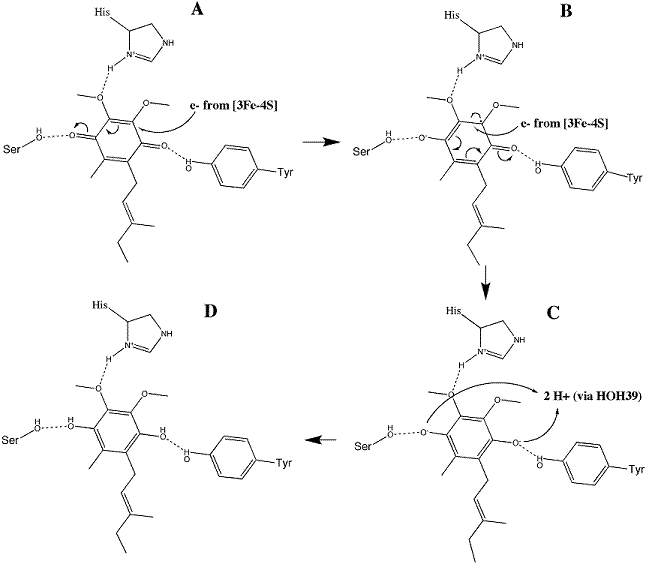
Ubiquinone is initially oriented in the active site such that the O1 carbonyl group interacts with Tyr83 of SdhD via hydrogen bonding. The electrons removed during the oxidation reaction are conveyed through the iron-sulfur clusters to 3Fe-4S. Their presence on that cluster stimulates a conformational change in the substrate, reorienting such that a second hydrogen bond between the of SdhC may form. The electrons are transferred to the substrate individually, where the addition of the first produces a radical semiquinone and addition of the second completes the reduction to ubiquinol. This mechanism is illustrated in image 3 [11].
Image 3: Reduction of ubiquinone to ubiquinol (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:QuinoneMechanism.gif).
Regulation
Since succinate dehydrogenase possesses multiple active sites that catalyze two different reactions, two main classes of competitive inhibitors function on the enzyme. The first class involves succinate analogs like the naturally-occurring TCA cycle intermediates malate and oxaloacetate, as well as synthetic analogs like malonate. Some of the strongest succinate dehydrogenase inhibitors act on the SdhA active site, most likely because it has only one site to block for its function to cease. The second class of inhibitors includes the ubiquinone analogs thenoyltrifluoroacetone and carboxin, which bind to the ubiquinone active site as electron acceptors, preventing reduction of the substrate via electrons traveling through the cytochrome[12].
References
- ↑ Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004 Mar 5;279(10):9424-31. Epub 2003 Dec 12. PMID:14672929 doi:10.1074/jbc.M311876200
- ↑ 2.0 2.1 Voet, Donald, Charlotte W. Pratt, and Judith G. Voet. Fundamentals of Biochemistry: Life at the Molecular Level. 2nd Ed. Hoboken, NJ: Wiley, 2008.
- ↑ Rutter J, Winge DR, Schiffman JD. Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion. 2010 June; 10(4):393-401
- ↑ Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001 Jul;69(1):49-54. Epub 2001 Jun 12. PMID:11404820 doi:10.1086/321282
- ↑ Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K. Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase). J Biochem. 2003 Aug;134(2):191-5. PMID:12966066
- ↑ 6.0 6.1 Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003 Jan 31;299(5607):700-4. PMID:12560550 doi:10.1126/science.1079605
- ↑ 7.0 7.1 Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006 Mar 17;281(11):7309-16. Epub 2005 Dec 27. PMID:16407191 doi:http://dx.doi.org/10.1074/jbc.M508173200
- ↑ Kenney WC. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975 Apr 25;250(8):3089-94. PMID:235539
- ↑ Boyd AW, Lackmann M. Signals from Eph and ephrin proteins: a developmental tool kit. Sci STKE. 2001 Dec 11;2001(112):re20. PMID:11741094 doi:10.1126/stke.2001.112.re20
- ↑ Vinogradov AD, Kotlyar AB, Burov VI, Belikova YO. Regulation of succinate dehydrogenase and tautomerization of oxaloacetate. Adv Enzyme Regul. 1989;28:271-80. PMID:2624174
- ↑ 11.0 11.1 11.2 Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200
- ↑ Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, Van Remmen H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem J. 2008 Jan 15;409(2):491-9. PMID:17916065 doi:10.1042/BJ20071162