User:Christina Evans/Sandbox 1
From Proteopedia
|
Description of Class I MHC
Class I major histocompatibility molecules are cell surface proteins that signal the presence of intracellular pathogens, such as viruses, to the host’s immune system. When viruses invade a host cell, they use the cell’s machinery to reproduce their nucleic acid and synthesize their proteins. Viral proteins, like cellular proteins, become targets for degradation and recycling, generating small portions of the viral proteins called peptides. Viral peptides of the right size can fit into the binding cleft of MHC-1 molecules, which are then displayed on the surface of the viral-infected cell. Cytotoxic T Lymphocytes (CTL) have receptors for foreign (viral) peptides complexed to MHC-1. When a CTL binds to a viral peptide-MHC-1 complex, the CTL secretes substances that lead to lysis and death of the infected cell, eventually clearing the infection. Thus, MHC-1 molecules function as an important warning system for the presence of intracellular infections. Viruses that develop a way to prevent expression of MHC-1 on the cell surface have a better chance of survival in the host.[1]
History of Class I MHC
Major Histocompatibility proteins are encoded by a diverse set of genes that mediate transplant acceptance and rejection. MHC genes were discovered by scientists working on tissue rejection in inbred strains of mice and rabbits. Two scientists, E.E. Tyzzer and Clarence C. Little were interested in the genetic differences that control the immune response to transplanted tissues in mice. In 1916, they transplanted tumors from different strains of mice, and reported the acceptance and rejection of grafts between different strains [2]. In a similar experiment, Bover found that cancer tissues were accepted if the donor and recipient were identical twins. Bover concluded that rejection and acceptance of transplanted tissues is genetically controlled. In the 1940s, Peter Medawar, while performing a tissue compatibility experiment with rabbits, confirmed that tissue graft rejection was triggered by an immune response. Medawar would perform skin grafts on rabbits to determine if the tissues were compatible, if the tissue was rejected he would transplant the same tissue again on the same recipient, and it was found that the tissue rejection occurred twice as fast. This indicated that the rejection was caused by an adaptive immune response, because the immune system was quicker to respond to the repeated procedure [3]. A few years later, George Snell ‘invented’ the congenic mouse strain. These mice were bred to be genetically identical, except at one locus, which is what made them congenic. During each new generation of mice, Snell would perform a skin allograft on them to determine if they would accept or reject the tissue. Snell would then continue to inbreed the mice that rejected the skin graft, but were identical in every other aspect. After many inbreeds, Snell was able to determine the location of tissue compatibility on the chromosome. Snell called the locus that controlled tissue compatibility, histocompatibility genes[4]. Later studies revealed several closely linked histocompatibility genes that are located on the same chromosome, and became known as the MHC gene complex. In humans, MHC proteins are also called the Human Leukocyte Antigen, or HLA, and can be used interchangeably. HLA was first discovered by Gorer in his work with blood antigens and tissue grafts. Gorer observed that recipients who received a tumor transplant from a donor that had different blood antigens, the tissue would be rejected. The genetic differences in the antigens present on blood cells would cause an immune reaction during a tissue donation, much like what occurs in mice when they have different MHC genes. The discovery of the HLA/MHC locus has prompted many studies in the field of tissue grafts, to help lower the risk of a patient receiving an incompatible tissue donation.
Stucture of Class I MHC
MHC-1 molecules are composed of two polypeptide or protein chains, an alpha ( chain and beta-2 microglobulin (. The α chain has three external domains designated α1, α2 and α3, a transmembrane region and a short cytoplasmic tail. The α1 and α2 domains interact to form the peptide-binding cleft composed of two (shown in indigo) regions and a bottom of antiparallel (shown in blue). β2M associates noncovalently with the α3 domain and stabilizes the complex[5]. Each chain is also stabilized by a located within the β sheets. The binding cleft can fit peptides 8-10 amino acids in length, and both viral and host cell peptides can be bound. The peptide is held in the by (shown in cyan) , which are amino acids located at the terminal ends of the peptide. Because the MHC-1 molecule only interacts with the terminal residues, it does not matter what the non-terminal residues are. Therefore, a single MHC-1 molecule can present many different viral peptides, as long as the terminal amino acids are correct for that MHC-1 molecule.
Each person has their own repertoire of MHC-I molecules, because they are highly polygenic and polymorphic. There are many genetic variants of MHC-I in the human population, this makes the MHC-I protein polymorphic. There are also many genes that code for MHC-I molecules, which makes MHC-I polygenic. We inherit our MHC-I molecules from our parents, both the mother and father contribute three α chains, so each person has six α chains total. The likelihood of an individual to inherit identical copies of their parent’s α chains is very low, because of the extremely varied, or polymorphic, nature of the alleles that control the expression of the α chain on MHC-I. This increased chance of a person being heterozygous for each MHC-I molecule is advantageous, because this ensures that a person will have plenty of different MHC-I molecules to protect them against many different viruses.
Formation of MHC-I Molecules
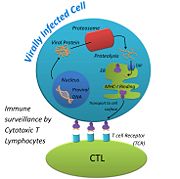
MHC-1 molecules are synthesized in ribosomes anchored in the membrane of the endoplasmic reticulum (ER). In the ER, the MHC-I molecules first associate with a chaperone protein called calnexin. This helps the very large MHC-I molecule fold correctly into place, and helps it retain a partially folded state in the ER. When Calnexin is released, β2M binds to the α chain on MHC-I. Chaperone proteins, calreticulin and tapasin, stabilize the MHC-1 configuration until a (shown in pink) is loaded into the binding cleft. Once the peptide is bound to MHC-I, Transporters Associated with Antigen Processing (TAP), tapasin, and calreticulin, dissociate from the MHC-1:peptide complex. This final, stable is targeted to the surface of the cell via the Golgi apparatus[6].
Formation of Viral Peptide
When a virus injects its nucleic acid into a host cell, it uses the cell’s machinery to replicate its genome and synthesize its proteins. Because the viral proteins are being made within the host cell, the viral proteins can undergo recycling just like host cell proteins. Misfolded proteins are marked for destruction in the cytosol by the addition of a small molecule called, ubiquitin. Once ubiquitin is attached, the protein is sent to the proteasome. Proteosomes are large cylindrical structures with many proteolytic enzymes. In the presence of a virus, the proteasome cleaves proteins into peptides 8-10 residues long with the appropriate anchor residues ensuring a stable fit into the peptide binding cleft of the MHC-I molecule. The peptides are transported from the cytosol to the lumen of the Endoplasmic Reticulum (ER) via ER membrane proteins, TAP.
Function of MHC-I
The function of MHC-I is to alert the immune system that something is wrong with the host cell[7]. When an infected cell displays a viral peptide via MHC-I, Cytotoxic T Lymphocytes (CTLs) are attracted to it. CTLs use their T Cell Receptor (TCR) to bind to the viral peptide that is on display. Each CTL has its own TCR, which is able to detect its own foreign peptide. This variability among TCRs is made possible when the genes of a T cell shuffle to form its own, unique TCR. This ensures that many different peptides can be recognized by CTLs, so our body is thoroughly protected. TCRs that bind to self-peptides that are displayed on MHC-I are destroyed before they are allowed to travel around the body[8]. Once a TCR finds an MHC-I displaying its complementary peptide, the CTL binds strongly to the peptide. The interaction is strengthened with the help of a glycoprotein called, Cluster of Differentiation 8 (CD8) that is found on the surface of the CTL. When the TCR binds the peptide, CD8 binds to the MHC-I complex, forming a stronger bond between the CTL and the host cell[9]. After a strong bond has been made, the CTL is activated by signaling pathways and it starts to release perforin, a protein that makes pores in the surface of the infected host cell. The pores help granzymes, proteases that are released by the CTL, enter the infected host cell and attack the cell from the inside. CTLs also release cytokines, proteins that are secreted by one cell and cause an effect on another, that help the CTLs multiply, and activates them so they will start attacking other infected host cells[10]. None of this can occur, however, if MHC-I is not able to perform its function, and display viral peptides on the host cells surface.
Effect of HCMV on MHC Class I
References
- ↑ 1.0 1.1 Gewurz BE, Gaudet R, Tortorella D, Wang EW, Ploegh HL, Wiley DC. Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6794-9. PMID:11391001 doi:http://dx.doi.org/10.1073/pnas.121172898
- ↑ Silverstein, Arthur. M. (1989). A History of Immunology. San Diego, California: Academic Press. Inc. pp.282-283.
- ↑ Ono, S. J. (2004). The birth of transplantation immunology: the Billingham-Medawar experiments at Birmingham University and University College London. The Journal of Experimental Biology. 207(23): 4013-4014.
- ↑ Penn, D. J. (2002). Major histocompatibility complex (MHC). Encyclopedia of Life Sciences.
- ↑ Becker, J.W. Reeke, Jr. G. N. (1985) Three-dimensional structure of β2-microglobulin. Immunology. 82:4225-4229.
- ↑ Goodsell, D. (2002). Molecule of the month: major histocompatibility complex. Protein Databank, 1-2. DOI: 10.2210/rcsb_pdb/mom_2005_2.
- ↑ Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. The major histocompatibility complex and its functions. Available from: http://www.ncbi.nlm.nih.gov/books/NBK27156/
- ↑ Goodsell, D. (2005). T-cell receptor. Protein Databank, 1-3. http://www.rcsb.org/pdb/education_discussion/molecule_of_the_month/download/T-Cell_Receptor.pdf.
- ↑ Varghese, J. C., Kane, K. P. (2008) TCR complex-activated CD8 adhesion function by human T cells. The Journal of Immunology. 181: 6002-6009.
- ↑ Irvine, D. J. (2003). Function-specific variations in the immunological synapses formed by cytotoxic T cells. PNAS. 100(24): 13739-13740.
- ↑ Chevalier, M.S., Daniels, G. M., Johnson, D. C. (2002) Binding of human cytomegalovirus US2 to major histocompatibility complex class I and II proteins is not sufficient for their degradation. Journal of Virology. 76(16): 8265-8275.
- ↑ Berg J.M., Tymoczko J.L., Stryer L. (2002). The Immunoglobulin Fold Consists of a Beta-Sandwich Framework with Hypervariable Loops. Biochemistry.
See Also

