User:Liz Thomas/Sandbox 1
From Proteopedia
|
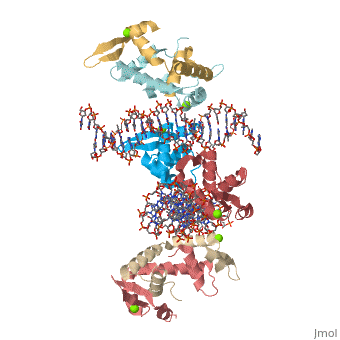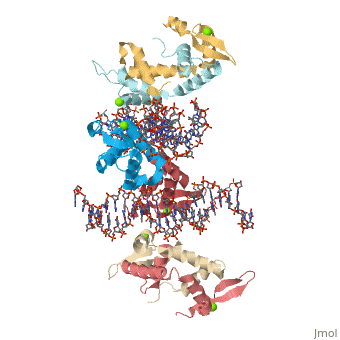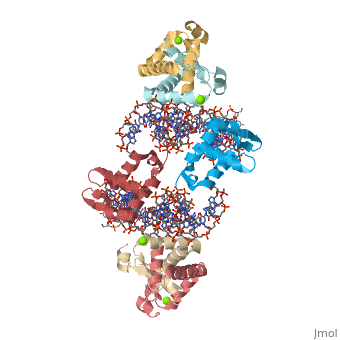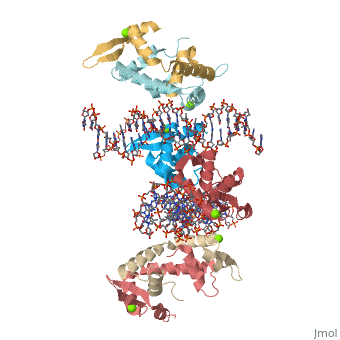
Crystal Structure of Foxp2 bound Specifically to DNA
New Article
|
| |||||||||
| 2a07, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | FOXP2, CAGH44, TNRC10 (Homo sapiens) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
FOXP2 is a transcription factor containing a winged-helix DNA binding domain, and is necessary for proper development of the lungs and brain. It is located on human chromosome 7 (7q31), and comprises 715 amino acids in its major splice form. FOXP2 is a member of the FOX (forkhead box) family and FOXP (FOXP1-4) subfamily of proteins, all of which contain a 90 amino acid winged-helix DNA binding motif. The foxP subfamily contains motifs like a glutamine-rich region, a leucine zipper, a zinc finger, and a forkhead domain, and is notable for conducting domain swapping.
Evolutionary Conservation
Because FOXP2 is the first protein to be solidly linked to a human speech disorder, much investigation has focused on its evolutionary history in hopes of elucidating the molecular mechanism behind the uniqueness of human speech. Evidence suggests that the protein underwent a recent selective sweep within the same time frame as the purported rise of language in modern humans, supporting this idea. Interestingly, molecular data also shows neandertals and modern humans share the same sequence.
Chimpanzee, gorilla, and rhesus monkey FOXP2 sequences are identical. The human version differs from these primate versions by two amino acid changes: a threonine to asparagine at position 303 and asparagine to serine at position 325. Researchers have hypothesized that the human-specific change at 325 creates a potential phosphorylation site by protein kinase C, with a predicted corresponding change in the protein's transcriptional regulation. Experiments in mouse and human neuronal cell models suggest that these human-specific changes play an important role in vocalization and neural function.
DNA Binding
Domain Swapping
Disease Mutations
Disease mutations in FOXP2 and related proteins correspond to either the domain-swapping dimer interface or the DNA binding sequence. FOXP2 and its family member FOXP3 are similar enough that the crystal structure of one can be used to explain the effects of mutation on structure in the other. In FOXP3, mutations at Ile363Val, Phe371Cys, Phe371Leu, Ala384Thr, and Arg397Trp have been traced to the autoimmune disorder [1]IPEX. With the exception of the Arg553His mutation, all of the following amino acid numbers in the titles refer to FOXP3.
Arginine to Histidine at 553
This is the only mutation that has been characterized in the original FOXP2 protein. Arg553 is a major component in the binding of FOXP2 to DNA. It creates a hydrogen bond with Thy11' and van der Waals contacts with Thy 11', Ade12', and Ade13'. A mutation to a histidine disrupts the interface between the protein and DNA, ostensibly preventing or reducing binding affinity.
Isoleucine to Valine at 363
Ile363 in FOXP3 corresponds to Ile530 in FOXP3. The Ile530Val mutation alters DNA binding by influencing neighboring residues that contact DNA. Ile530 (H2) creates van der Waals contacts with Leu527, Leu556, and Trp 573. Leu527 and Trp573 in turn directly contact the DNA backbone, while Leu556 holds helix 3 in place to facilitate DNA recognition. The removal of a single methyl group when isoleucine is changed to valine destabilizes the entire binding interface.
Alanine to Threonine at 384
Ala384 in FOXP3 correponds to Ala551 in FOXP2. Mutation of alanine to threonine at position 384 (H3) introduces an extra methyl group, causing steric interference in the restrictive protein/DNA juncture and disrupting DNA binding.
Arginine to Tryptophan at 397
Arginine 397 in FOXP3 corresponds to Arg564 in FOXP2. Arg564 (S2) binds moieties on the DNA backbone and in the minor groove. Mutation of Arg564 to a bulky tryptophan destabilizes protein/DNA interaction through steric hindrance.
Phenylalanine to Cystine at 371 and Phenylalanine to Leucine at 371
Phe371 in FOXP3 corresponds to Phe538 in FOXP2. Phe538 is not located near the DNA binding face, and so probably plays little to no role in direct DNA binding. It is, however, at the center of the domain-swapped dimer interface. Mutations at this Phe may then cause disease by disturbing the core of the domain-swapped dimer.
References
Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. 2006. Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14: 159-166. doi:10.1016/j.str.2005.10.005
Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413: 519-523. doi:10.1038/35097076 PMID 11586359.
Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin JJ, Hanni C, Fortea J, de la Rasilla M, Bertrapetit J, Rosas A, Paabo S. 2007. The derived FOXP2 variant of modern humans was shared with neandertals. Current Biology 17:1-5. doi:10.1016/j.cub2007.10.2008
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006 Jan;14(1):159-66. PMID:16407075 doi:10.1016/j.str.2005.10.005
Categories: Homo sapiens | Bates, D L. | Chen, L. | Han, A. | Nowick, K. | Paabo, S. | Stroud, J C. | Tong, H. | Wu, Y. | Double-helix | Forkhead | Homodimer | Magnesium | Monomer | Swapping | Winged-helix