User:Stéphanie Kraemer/Sandbox 106
From Proteopedia
p53R2
| |||||||
| 3hf1, resolution 2.60Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | RRM2B, P53R2 (Homo sapiens) | ||||||
| Activity: | Ribonucleoside-diphosphate reductase, with EC number 1.17.4.1 | ||||||
| Related: | 1xsm, 2uw2, 1w69, 1w68 | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
P53R2 is an oxydoreductase composed of 351 residues. It is a small subunit of the ribonucleotide reductase (RNR). RNR catalyses the reduction of the four nucleotides to desoxyribonucleotides. It exists three classes of RNR. Class I RNR is a tetramer composed of the two types of subunits with stoichiometry α2β2 and three subunits have been identified in mammals :
- Large (α) subunit M1 that contains the enzyme active site
- Small (β) subunit M2 that contains a dinuclear iron site, it’s the regulatory subunit
- P53R2, the last identified (in 2000), which is transactivated by p53 in response to DNA damage in cells during the G0-G1 cell cycle phase
M2 and p53R2 interact with M1 through the C-terminal binding domain. These two subunits share more than 80% sequence identity. But the few differences between the two are not unimportant, as it’s explained below. The first X-ray crystal structure of p53R2 has a resolution of 2,6 Å and permits to describe its structure and also to show the structural differences with the M2 subunit.
Structure and function
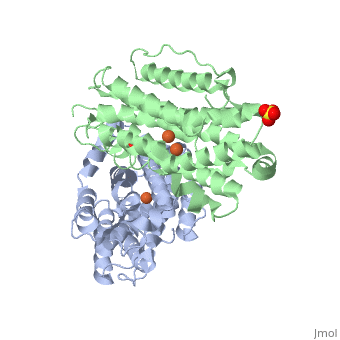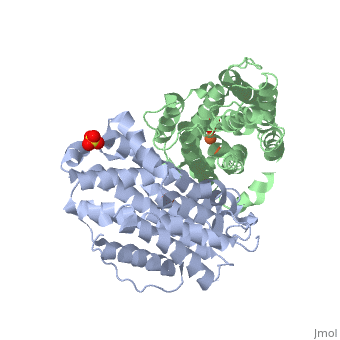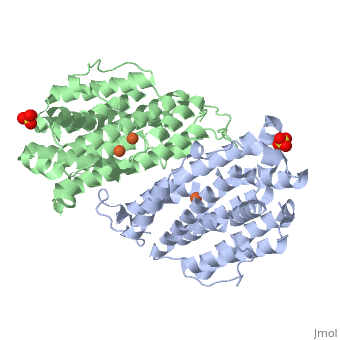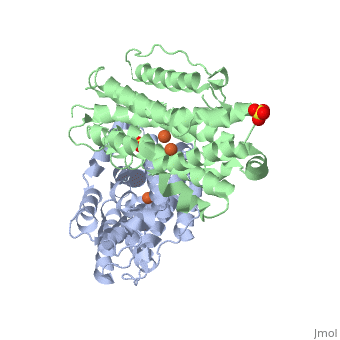
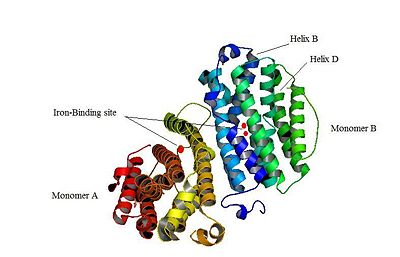
The X-ray crystal structure permits to see that p53R2 is made of two monomers A and B themselves made of loops and helix. Two of them play an important role. is highlighted. But concerning this site, the two monomers are not the same. Actually, the B monomer has two iron-binding site (called Fe2 and Fe1) whereas the A monomer has only one which is Fe2. This can be explained by structural changes in the helix that compose the two monomers. The 37 to 42 N-terminal residues (called the swivel loop) from one monomer can rotate between two conformations and can influence the position of the helix B or D on the opposite monomer.
The N-terminal residues of the monomer A can stabilize the B helix of the monomer B due to different interactions. R41 of the monomer A forms a salt bridge with E119 of monomer A. This interaction permits the formation of a H-bond between R40 of monomer A with G101of monomer B. Furthermore K37 in monomer A forms a salt bridge with E105 of monomer B and this stabilize its B helix. All the interactions allow D100 of monomer B to be well oriented to bind Fe1 (see Figure 1 from On the contrary R40 of monomer B is bound to E119 of the same monomer and so it can not bind to G101 of the monomer A and the consequence is that F42 disturb the B helix of monomer A. D100 can not interact with Fe1. This explain why the monomer A has only one iron-binding site whereas the monomer B has two. Compare to the M2 subunit, these iron-binding sites are less efficient. This is due to the different conformations that the p53R2 subunit can adopt (stabilization or not of the two helix B and D). Furthermore the iron is the cofactor of the reaction catalyses by the RNR. The fact that in the p53R2 subunit the iron-binding is less efficient permits to imagine a specific anti-cancer therapy that targets these region, for example the drug deferoxamine mesylate an iron chelator. Without iron, the reduction of the nucleotides can not take place and this could avoid the proliferation of cancer cells.