We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
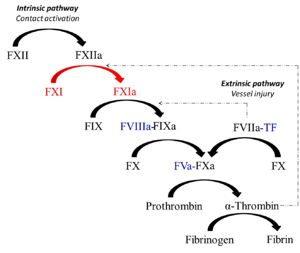
Factor XIa
From Proteopedia
(Redirected from Factor XI)
| |||||||||||
References
- ↑ Thompson RE, Mandle R Jr, Kaplan AP. Association of factor XI and high molecular weight kininogen in human plasma. J Clin Invest. 1977 Dec;60(6):1376-80. PMID:915004 doi:http://dx.doi.org/10.1172/JCI108898
- ↑ Baglia FA, Gailani D, Lopez JA, Walsh PN. Identification of a binding site for glycoprotein Ibalpha in the Apple 3 domain of factor XI. J Biol Chem. 2004 Oct 29;279(44):45470-6. Epub 2004 Aug 17. PMID:15317813 doi:10.1074/jbc.M406727200
- ↑ Herwald H, Jahnen-Dechent W, Alla SA, Hock J, Bouma BN, Muller-Esterl W. Mapping of the high molecular weight kininogen binding site of prekallikrein. Evidence for a discontinuous epitope formed by distinct segments of the prekallikrein heavy chain. J Biol Chem. 1993 Jul 5;268(19):14527-35. PMID:7686159
- ↑ Bouma BN, Griffin JH. Human blood coagulation factor XI. Purification, properties, and mechanism of activation by activated factor XII. J Biol Chem. 1977 Sep 25;252(18):6432-7. PMID:893417
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
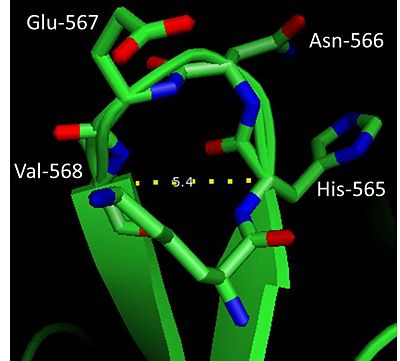
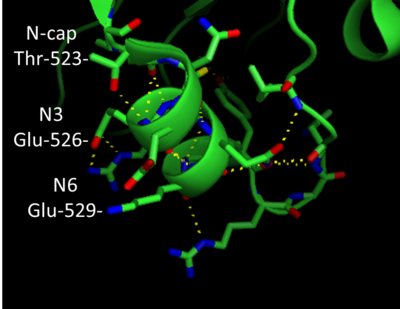
- ↑ Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994 Dec;3(12):2207-16. PMID:7756980 doi:http://dx.doi.org/10.1002/pro.5560031206
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Dorfman R, Walsh PN. Noncovalent interactions of the Apple 4 domain that mediate coagulation factor XI homodimerization. J Biol Chem. 2001 Mar 2;276(9):6429-38. Epub 2000 Nov 22. PMID:11092900 doi:10.1074/jbc.M010340200
- ↑ McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human coagulation factor XI: the presence of tandem apple domains. Biochemistry. 1991 Feb 26;30(8):2056-60. PMID:1998667
- ↑ Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 2008 Jan;24(1):161-6. PMID:18187866
- ↑ Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009 Feb;8(2):651-61. PMID:19159218 doi:10.1021/pr8008012
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003 Oct 2;425(6957):535-9. PMID:14523451 doi:10.1038/nature01962
- ↑ Sinha D, Marcinkiewicz M, Navaneetham D, Walsh PN. Macromolecular substrate-binding exosites on both the heavy and light chains of factor XIa mediate the formation of the Michaelis complex required for factor IX-activation. Biochemistry. 2007 Aug 28;46(34):9830-9. Epub 2007 Aug 4. PMID:17676929 doi:10.1021/bi062296c
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
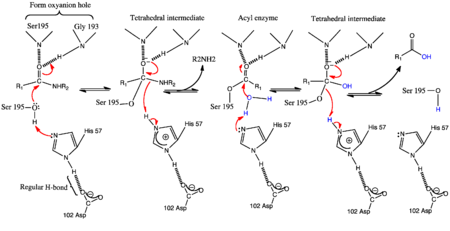
- ↑ Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331-58. PMID:332063 doi:http://dx.doi.org/10.1146/annurev.bi.46.070177.001555
- ↑ Bender ML, Killheffer JV. Chymotrypsins. CRC Crit Rev Biochem. 1973 Apr;1(2):149-99. PMID:4372018
- ↑ Blow DM, Birktoft JJ, Hartley BS. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337-40. PMID:5764436
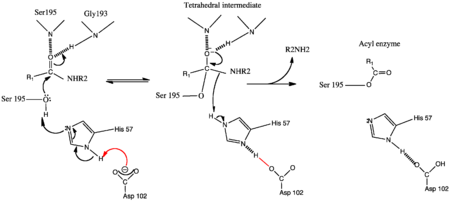
- ↑ Kossiakoff AA, Spencer SA. Direct determination of the protonation states of aspartic acid-102 and histidine-57 in the tetrahedral intermediate of the serine proteases: neutron structure of trypsin. Biochemistry. 1981 Oct 27;20(22):6462-74. PMID:7030393
- ↑ Markley JL, Ibanez IB. Zymogen activation in serine proteinases. Proton magnetic resonance pH titration studies of the two histidines of bovine chymotrypsinogen A and chymotrypsin Aalpha. Biochemistry. 1978 Oct 31;17(22):4627-40. PMID:31898
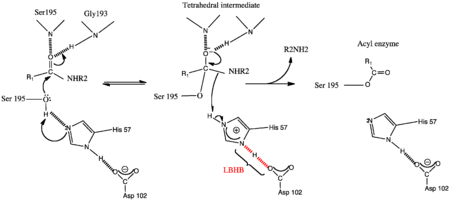
- ↑ Frey PA, Whitt SA, Tobin JB. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994 Jun 24;264(5167):1927-30. PMID:7661899
- ↑ Frey PA, Whitt SA, Tobin JB. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994 Jun 24;264(5167):1927-30. PMID:7661899
- ↑ Henderson R. Catalytic activity of -chymotrypsin in which histidine-57 has been methylated. Biochem J. 1971 Aug;124(1):13-8. PMID:5126468
- ↑ Craik CS, Roczniak S, Largman C, Rutter WJ. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987 Aug 21;237(4817):909-13. PMID:3303334
- ↑ Kidd RD, Sears P, Huang DH, Witte K, Wong CH, Farber GK. Breaking the low barrier hydrogen bond in a serine protease. Protein Sci. 1999 Feb;8(2):410-7. PMID:10048334
- ↑ Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986 Nov 18;25(23):7751-9. PMID:3542033
- ↑ Kidd RD, Sears P, Huang DH, Witte K, Wong CH, Farber GK. Breaking the low barrier hydrogen bond in a serine protease. Protein Sci. 1999 Feb;8(2):410-7. PMID:10048334
- ↑ Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986 Nov 18;25(23):7751-9. PMID:3542033
- ↑ Brady K, Wei AZ, Ringe D, Abeles RH. Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors. Biochemistry. 1990 Aug 21;29(33):7600-7. PMID:2271520
- ↑ Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001 Jan 30;40(4):853-60. PMID:11170405
- ↑ Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008 Nov;6(11):1876-83. Epub 2008 Aug 28. PMID:18761718 doi:10.1111/j.1538-7836.2008.03143.x
- ↑ McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human plasma prekallikrein: the presence of four novel apple domains in the amino-terminal portion of the molecule. Biochemistry. 1991 Feb 26;30(8):2050-6. PMID:1998666
- ↑ Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008 Nov;6(11):1876-83. Epub 2008 Aug 28. PMID:18761718 doi:10.1111/j.1538-7836.2008.03143.x
- ↑ Saito H, Ratnoff OD, Bouma BN, Seligsohn U. Failure to detect variant (CRM+) plasma thromboplastin antecedent (factor XI) molecules in hereditary plasma thromboplastin antecedent deficiency: a study of 125 patients of several ethnic backgrounds. J Lab Clin Med. 1985 Dec;106(6):718-22. PMID:4067382
- ↑ 10706899
- ↑ Wang X, Cheng Q, Xu L, Feuerstein GZ, Hsu MY, Smith PL, Seiffert DA, Schumacher WA, Ogletree ML, Gailani D. Effects of factor IX or factor XI deficiency on ferric chloride-induced carotid artery occlusion in mice. J Thromb Haemost. 2005 Apr;3(4):695-702. Epub 2005 Feb 23. PMID:15733058 doi:10.1111/j.1538-7836.2005.01236.x
- ↑ Riley PW, Cheng H, Samuel D, Roder H, Walsh PN. Dimer dissociation and unfolding mechanism of coagulation factor XI apple 4 domain: spectroscopic and mutational analysis. J Mol Biol. 2007 Mar 23;367(2):558-73. Epub 2006 Dec 29. PMID:17257616 doi:10.1016/j.jmb.2006.12.066
- ↑ Kravtsov DV, Wu W, Meijers JC, Sun MF, Blinder MA, Dang TP, Wang H, Gailani D. Dominant factor XI deficiency caused by mutations in the factor XI catalytic domain. Blood. 2004 Jul 1;104(1):128-34. Epub 2004 Mar 16. PMID:15026311 doi:10.1182/blood-2003-10-3530
- ↑ Kravtsov DV, Wu W, Meijers JC, Sun MF, Blinder MA, Dang TP, Wang H, Gailani D. Dominant factor XI deficiency caused by mutations in the factor XI catalytic domain. Blood. 2004 Jul 1;104(1):128-34. Epub 2004 Mar 16. PMID:15026311 doi:10.1182/blood-2003-10-3530