Image:Positions of the mutations in PR variants used for structural studies.jpg
From Proteopedia
Size of this preview: 625 × 599 pixels
Full resolution (652 × 625 pixel, file size: 98 KB, MIME type: image/jpeg)
Summary
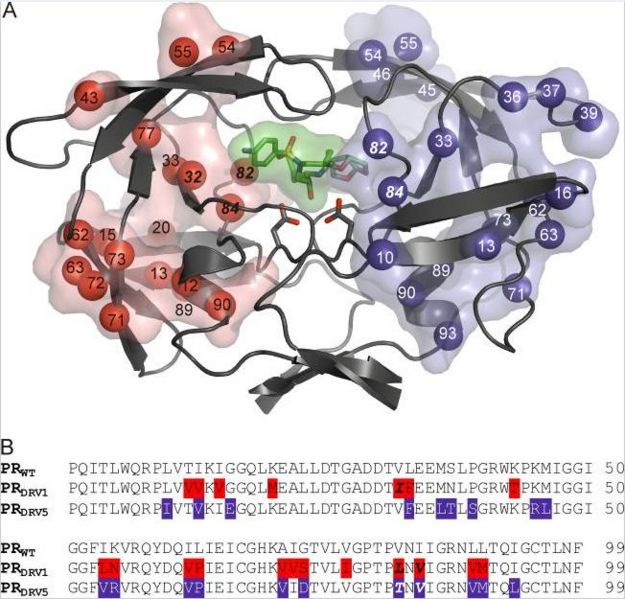
Positions of the mutations in PR variants used for structural studies. (A) Each monomer of PR is used to depict the mutations in the PRDRV1 and PRDRV5 variants in red and blue, respectively. Mutated residues are represented with their Cα atoms (spheres) and their transparent solvent-accessible surfaces. Active-site aspartates are shown as sticks; darunavir bound to the active site is represented by sticks, and its solvent-accessible surface area is green. (B) Sequence alignment of PRDRV1 and PRDRV5 with the wild-type (WT) sequence. Primary mutations in residues interacting directly with darunavir are in boldface italics.
Licensing
{{subst:Permission from license selector}}
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | User | Dimensions | File size | Comment | |
|---|---|---|---|---|---|
| (current) | 18:27, 29 December 2012 | Angelika Wackerl (Talk | contribs) | 652×625 | 98 KB | Positions of the mutations in PR variants used for structural studies. (A) Each monomer of PR is used to depict the mutations in the PRDRV1 and PRDRV5 variants in red and blue, respectively. Mutated residues are represented with their Cα atoms (spheres) |
- Edit this file using an external application
See the setup instructions for more information.
Links
The following pages link to this file:
Metadata
This file contains additional information, probably added from the digital camera or scanner used to create or digitize it. If the file has been modified from its original state, some details may not fully reflect the modified image.
| Date and time of data generation | 18:18, 29 December 2012 |
|---|---|
| Author | Angelika Wackerl |
| Date and time of digitizing | 18:18, 29 December 2012 |
| DateTimeOriginal subseconds | 06 |
| DateTimeDigitized subseconds | 06 |
