Sandbox89220
From Proteopedia
Contents |
Phosphomannose mutase 2
Background
Structure
|
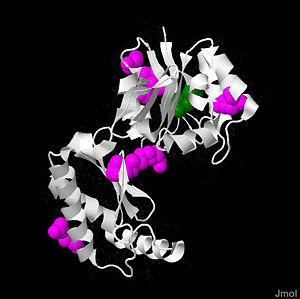
PMM2 is a 246 amino acid long protein, with 2 identical subunits having close resemblance in their working as independent domains, hence it’s also known as a homodimeric protein. Even if the structure of PMM2 is already solved,however, it's still not clear where its domains and its critical motifs are. Nonetheless, through the extraploration of PMM1, it’s known to be classified as the member of HAD (haloacid dehydrolase) superfamily due to the fact that it had 4 different types of motifs[10], each of which is highly conserved across the vertebrates’ lineages during evolutions. Similar to other members in HAD superfamily, the two domains are separately recognized as core and cap domain. The core domain (residues 1 – 90, and 198-262)of PMM2 displays the characteristic 4 motifs of HAD superfamily that contribute to the catalytic ability of the active sites in the protein. Motif 1 has as nucleophile which serves as the mediator for the phosphoryl group transfer, the second Asp, on the other hand, acts as in the general acid-base reaction. While motif 1 provides the nucleophilic and general acid-base reactions, motif 2 in the protein which usually contains or helps in positioning or binding on the substrates’ phosphoryl group. In addition, motif 3 has the typical Lys or Arg residues while motif 4 has the acidic residues (Asp and Glu) that bind magnesium cofactor[11][2]. All of the residues that play the critical role in catalytic activity of the protein are well throughout the evolutionary path. The cap domain of PMM2 is smaller, functioning as the regulator for the access of the substrate into the active site of the core domain and at the same time contains the primary and secondary substrate specificity loops that recognize only specific substrate. It's still unclear how the PMM2 would work together to catalyze the transfer of phosphoryl group within its substrate, but it's possible that it might have the same machinery as the PGM(phosphoglycerate mutase) protein family.
How does PMM2 work?
PMM2 converts mannose-6-phosphate into mannose-1-phosphate that would then serve as the building bloc of the dolichol-linked oligosaccharides for glycosylation in endoplasmic reticulum. It's not clear how PMM2 the cap and core domains would coordinate with each other. However,through the extrapolation of its paralogous protein, PMM1 which has a complex mechanism that uses charges of the conserved residues in the cap domains to sweep the substrate into core domain, we could postulate that PMM2 would utilize similar machinery[3]. Also, with the close resemblance to the PGM proteins, PMM2 might have similar machinery that manipulate the conformational changes that “closes” cap domain after binding substrates to allow the specificity loops getting into contact with the active site of the core domain, thus participating in catalysis[12].
Below shows the conversion of M6P into M1P(in cytosol):
PMM2,Glycans, and CDG
Imagine that endoplasmic reticulum in a cell as a big and busy protein synthesis site where a lot of modifications and different "accessories" are added in response to different signal carried by the proteins. To think about what are glycans, they are simply oligosaccarides or polysaccarides(the accessories)attached distinctively to the proteins that could give the proteins their characteristic functions or allow them to fold properly and funcion. There are 5 different types of glycans [13] synthesized in the body to aid in different biochemical reactions, namely the N-linked glycans, O-linked glycans, phosphoglycosyl proteins, C-mannosyl proteins, and Glypiated-linked proteins[4]. Each of the carbohydrate-peptide linkage contributes differently in proteins, enhancing their functions in cells. There are eleven different human CDG types result from the defects in the N-glycosylation pathway while 3 types are due to defects in O-glycosylation pathway, each has different phenotypes in patients. Amongst the defects in the N-glycosylation pathway, defects in PMM2 give rise to a more serious clinical implications in the patients, looking at its role in synthesizing building bloc of dolichol-linked oligosaccarides for N-glycans in endoplasmic reticulum, the precursor for all "high-mannose"(so-called due to the fact that it has higher content of mannose) glycans. Keeping in mind that ER as a big factory processing the "accessories" and attach them nicely onto the targeted proteins, we can easily see the impacts on the progression of producing the "accessorries"(the dolichol-linked glycans) if the main switch towards all the working machines is defective, in this case, the PMM2 as the switch to all.With the mutations in the PMM2 genes, the catalytic activities at the active sites of the expressed protein will then decrease, giving rise to low conversion of mannose-6-phosphate to mannose-1-phosphate (now as the raw materials to start all the processing in the factoty), which results in little substrate for the assembly of oligosaccharides in the subsequent bonding of the glycoconjugates in the ER lumen. Hence, hypoglycosylation of the proteins is the result of the defects in PMM2.
Evolutionary History
Both PMM1 and PMM2 are grouped under the haloacid dehydrogenase family, which has 4 characteristic motifs that are highly conserved in both PMMs. The phylegenic analysis of PMMs in 26 species of animals reveals a good complementary data to explain the divergence of the functions between these so closely related proteins in humans. Apparently, in the early stages of evolution of vertebrates or before the divergence of vertebrates, there are the duplication events of the PMM gene in the common vertebrates’ ancestor. The duplication allows extra set of genomic information to be passed on and mutations to occur, which later leads to the emergence of the similar yet distinct PMM1 and PMM2[2]. Judging from the higher degree of identity between certain yeast type and human’s PMM2,it's said that PMM2 evolves more slowly than PMM1. As a matter of fact, with the different evolutionary rates after duplication, it allows the PMM1 and PMM2 to have diverged functions in their active sites which have some degree of conservations throughout the evolutionary lineage. It doesn't escape the attention that there are different residues replacement in primary and secondary specificity loop in the highly conserved motifs 1 and 2 in both proteins. For example, while PMM1 can be stimulated by IMP in the brain to increase its phosphatase activity (as glucose-1,6- bisphosphatase) and has decreased activity as phosphomutase by then, IMP is shown to has no effect on both the phosphatase and phosphomutase activity of PMM2. Again, it’s obvious that the evolutionary development of the PMM genes after duplication results in these two similar yet distinct proteins.clinical manifestation
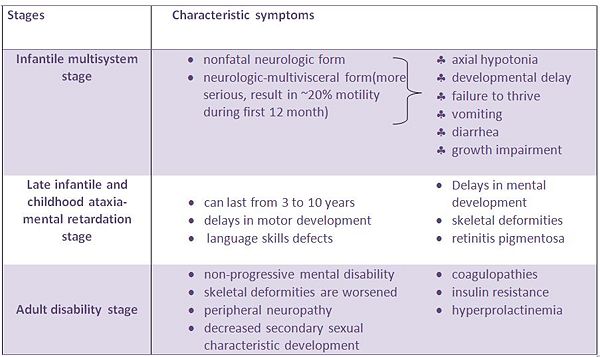
PMM2 is very critical during the early development of embryo, it's shown in experiment that the disruption of the genes causes the embryonic lethality. In addition, it's reported that wide spectrum of clinical manifestations is due to different mutation sites on the PMM2 genes, certain mutations are demographically specific, however, the most common mutation is the R141H amongst the patients from all around the world[5]. The patients will show psychomotoric retardation, muscle hypotonia, abnormal eye movements, inverted nipples, abnormal adipose tissue distribution, and slightly enlarged liver[6].The table below show CDG symptoms for different stages:
Leishmaniasis and PMM2
PMM2 is not only important in survival and development in humans, it’s found that there is another form of PMM2 that’s expressed in the protozoan parasite Leishmania Mexicana[14]which is believed to contribute to the virulence of the protozoan and enable them to survive in human host[7]. Since leishmaniasis[15] is prevalent world-wide, finding an inhibitor to get rid of its virulence in hosts is very promising when it’s shown that the PMM in this particular protozoan has a close proximity to the structure of PMMs in human beings. The PMM in Leishmania Maxicana has about the same function as that of PMMs in humans, it’s crucial in synthesizing the activated mannose block for glycoconjugates for the survival of the parasites in host. However, as mentioned by Kedzierski et al., 2006, despite the fact that the close resemblance of the PMM in Leishmania Maxicana to PMMs in human can be an exciting discovery to thrive the pharmaceutical field for the drugs to kill the parasites, it also poses problems when the drugs could not distinguish between humans’ PMMs from PMM in the parasites.References
- ↑ Chung et al,Domain motion and interdomain hot spots in a multidomain enzyme[1]
- ↑ 2.0 2.1 Rita Quental et al., Evolutionary History and Functional Diversification of Phosphomannomutase Genes[2]
- ↑ Freeze,TOWARDS A THERAPY FOR PHOSPHOMANNOMUTASE 2 DEFECIENCY, THE DEFECT IN CDG-Ia PATIENTS [3]
- ↑ Benjamin G. Davis, Synthesis of Glycoproteins[4]
- ↑ Quelhas et al.,Congenital Disorder of Glycosylation Type Ia: Searching for the Origin of Common Mutations in PMM2[5]
- ↑ Presentation of congenital disorders of glycosylation type 1a[6]
- ↑ George N. Phillips Jr et al., Structures of proteins of biomedical interest from the Center for Eukaryotic Structural Genomics[7]