Sandbox 171
From Proteopedia
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. | |||||||||
| |||||||||
| 2mys, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Overview
Myosin is one of three major classes of molecular motor proteins: myosin, dynein, and kinesin. As the most abundant of these proteins myosin plays a structural and enzymatic role in muscle contraction and intracellular motility. Myosin was first discovered in muscle in the 19th century. [1]
Crystallization and X-ray diffraction
Myosin is found in abundance, therefore it can be prepared in gram quantities. [2] For nearly 30 years the myosin head was resistant to crystallization, yet by 1993 researchers discovered a mechanism to obtain x-ray quality crystals. [2] The process modified the protein by reductive methylation. [2] X-ray data was used to determine the tertiary structure of the protein. [2]
Structure
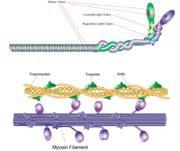
Myosin has a molecular size of approximately 520 kilodaltons with a total of six subunits. It has two 220 kD heavy chains which make the majority of the overall structure and two pairs of light chains which vary in size.[2] The molecule is asymmetric, having a long tail and two globular heads. [2] Each heavy chains composes the bulk of one of the globular heads. [2] Subfragment-1(S1) also termed the myosin head consists of ATP, actin, and two light chain binding sites.[2] Each globular head has a heavy chain and two light chains for a combined molecular size of about 130 kD. [2]
The myosin head is assymetrical with a length of 165 Angstroms and 65 Angstroms in width, with a total thickness of about 40 Angstroms. [2] About 48% of the amino acid residues in the myosin head are dominated by α helices. [2] At the carboxyl terminus one long α helix of about 85 Angstroms extends in a left-handed coil. [2] This particular helix forms the light chain binding region of the globular domain [2] The amino terminus of each heavy chain has a large globular domain containing the site of ATP hydrolysis.
Function
Molecules of myosin aggregate in muscle cells to form thick filaments. [3] The rodlike structure of these thick filaments act as the core in the muscle contractile unit. [3] The aggregation of several hundred myosin forms a bipolar structure which stacks in regular arrays. Muscles consist of another protein called actin. Actin forms the thin filament in muscle fibers. Myosin and actin interact through weak bonds. Without ATP bound, the myosin head binds tightly to actin. [3] With ATP bound, myosin releases the actin subunit and interacts with another subunit further down the thin filament. [3] This process continues in cycle, producing movement. Interaction of myosin and actin is regulated by two other proteins, tropomyosin and troponin. [3]
The cycle of myosin-actin interaction is outlined as follows: [3]
1. ATP binds to myosin and a binding site opens on myosin head to disrupt the actin-myosin interaction, actin is released. ATP is hydrolyzed
2. a conformational change moving the protein to a "high-energy" state causes the myosin head to change orientation moving it to bind with the actin subunit closer the a region called the Z disk than the previous actin subunit
3. the binding site is closed, strengthening the myosin-actin binding
4. a power stroke quickly follows and the myosin head undergoes an additional conformational change bringing it back to the resting state in which it began
Click the link to access DNAtube video "A Moving Myosin Motor Protein" http://www.dnatube.com/video/389/A-Moving-Myosin-Motor-Protein-myosin-actin-interaction
Literature Cited
- ↑ Spudich JA, Finer J, Simmons B, Ruppel K, Patterson B, Uyeda T. Myosin structure and function. Cold Spring Harb Symp Quant Biol. 1995;60:783-91. PMID:8824453
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50-8. PMID:8316857
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Nelson, D. and Cox, M.(2005). Lehninger Principles of Biochemistry. 4th ed. p.1119.