Sandbox 177
From Proteopedia
| |||||||||
| 3es9, resolution 3.40Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | CYPOR, Por (Rattus norvegicus) | ||||||||
| Activity: | NADPH--hemoprotein reductase, with EC number 1.6.2.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
NADPH-cytochrome P450 oxidoreductase
General Information
Horecker first identified this protein in 1950 as NADPH-specific cytochrome c reductase, based on his assumption that it was the redox partner for cytochrome c, found in the mitochondria.[1] However, studies in the 1960s and later showed that its main function is actually as the redox partner for cytochrome P450 in microsomal electron transport chains, resulting in a name change.[2][3] Today this protein is known as NADPH-cytochrome P450 oxidoreductase (CYPOR).
CYPOR is a ~78kDa, multidomain flavoprotein.[2] Containing three co-factors, FAD, FMN and NADPH, this protein is the archetype for the mammalian diflavin-containing enzyme family.[2] Research indicates that the protein, and other FAD/FMN binding proteins, are likely the product of the fusion of two ancestral genes.[4] This would account for the two distinct binding domain areas, FMN and FAD/NADPH, which each provide different functional capabilities to the overall protein.[4]
Regulation of this protein, which is found all tissues to some extent, is largely at the transcriptional level.[5] The thyroid hormone T3 acts as a hormonal regulator in most cases, while adrenocorticotrophic hormone acts as a regulator in a few specific cases.[6][7]
Structure
|
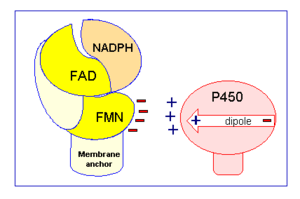
is a complex, multidomain protein composed of three asymmetric chains, (, , ). It also has three different types of associated ligands; one , three and two (Fig 1).[2] The three associated binding domains for these ligand types, a connecting domain and a transmembrane anchor make up the important structural elements of CYPOR (Fig 2).
The N-terminus of CYPOR consists of a single alpha-helix composed of 25 amino acids that functions as a transmembrane anchor (~6kDa), holding the protein in the endoplasmic reticulum.[2][8] The remaining, soluble ~66kDa portion of the protein, responsible for reducing cytochrome P450, consists of three binding domains for the ligands involved in the electron transport chain.[2] The FMN binding domain is composed of the first 170 residues of the soluble region, which are very similar to those of flavodoxin, another FMN binding protein.[2] The FAD and NADPH binding domains are located closer to the C-terminus, and are very similar to the FAD domain in ferredoxin-NADP+ oxidoreductase, both in terms of sequence and structure.[2]
Between the FMN and FAD/NADPH bind domains is a connecting domain, which is a highly flexible random coil.[2] The hinge region is composed of 12 residues from , and is highly conserved among most known CYPOR proteins, including those found in humans, rats and even yeast.[2] For simplicity the hinge region has only been highlighted in the A chain (Fig 1), however it is present in all three chains. The hinge section is presumed to be responsible for the relatively increased mobility of the FMN domain, changes to conformation and the relative orientation of the binding domains.[2] Studies that examined the rate of electron transfer within CYPOR seem to confirm this, as electron transfer rate appears to decrease proportionally to increases in the viscosity of the fluid medium it is in.[2] Within the hinge, if residues are mutated or added (2-4 residues), the effect on electron transport is positive or negligible.[2] However the same study found that the removal four residues from the hinge, as seen in Figure 1, prevents CYPOR from effectively transferring electrons to cytochrome P450, unless there is a high electron pool available.[2] This indicates that without the hinge movement electrons are not able to be efficiently moved from FAD to FMN, decreasing the reductase capabilities of CYPOR.[2]
Function
In vivo CYPOR is believe to alternate between a one and a three electron reduced form. While the 1 electron form is fairly stable, forming a neutral blue semiquinone, it is the hydroquinone, or 3 electron form, that is able to donate electrons to the desired redox partners.
As part of the microsomal electron transport system, CYPOR moves electrons from:
- NADPH → FAD → FMN → Cytochrome P450
Specifically a hydride anion is moved from NADPH to the FAD.[2] The two electrons are then individually passed to FMN, in a process that is believed to be conformationally gated.[2] As previously discussed in the structure section, this belief is based upon the fact that electrons do not appear to be able to be transferred from FAD to FMN unless the two ligands are in close proximity.[2] The protein accomplishes this by undergoing a conformational change, believed to occur because of the flexibility of the hinge domain, which brings the flavin isoalloxazine rings of FMN and FAD into close proximity to one another.[2] In this closed conformation van der Waals forces help hold the ligands together, allowing for efficient electron movement.[2] However, for CYPOR to transfer the electrons, again one at a time, from FMN to cytochrome P450, CYPOR cannot be in a closed conformation because it prevents cytochrome P450 from being able to access FMN.[2] As a result CYPOR must undergo another conformational change so it is in an open conformation allowing the necessary surface residues on the FMN binding domain to form interactions with cytochrome P450 for electron transfer to occur (Fig 2).[2] These surface residues have been found to have an increased number of carboxyl containing amino acids (Aspartate and glutamate), which gives this area a negative charge (Fig 2).[9][10] The carboxyl groups can then bind basic residues like leucine found on cytochrome P450 to correctly orient the two proteins for electron transfer.[9][10] Additionally, cytochrome P450 can have an induced dipole moment across it, with a partial positive charge occurring on the side of the protein where the internal heme is closest to the surface of the protein (Fig 2). [9][10] The partial positive charge interacting with the previously mentionned acidic residues of CYPOR is thought to help solidify the interaction between CYPOR and cytochrome P450 in the best orientation for electron transfer.[9][10]
This reduction of cytochrome P450 allows it to function in biosynthesis and biodegradation pathways of a variety of endogenous and foreign hydrophobic substrates, including drugs and steroids.[2][11] Cytochrome b5, cytochrome c and heme oxygenase can also receive electrons from CYPOR.[2] In these cases CYPOR is functioning in the heme degradation pathway, or with monooxygenase and/or 7-dehydrocholesterol reductase in sterol synthesis.[2]
Medical Significance
Cancer - Studies have shown that the reductase activity of CYPOR is capable of activating anticancer prodrugs.[2][12] Elevated expression of CYPOR has been found to increase the sensitivity of cancerous cells to certain anticancer drugs like tirapazamine.[12] This makes it a potential target for anticancer research and therapy.
Embryology & Development - CYPOR is believed to play a key role in the spatial and temporal expression of various signaling factors that are key in establishing correct embryogenesis and development pathways.[13] Studies with mice have shown that CYPOR is critical for mice embryos to progress into and past mid-gestation, as embryos lacking both CYPOR alleles did not survive past day 13.5 of gestation. [13] Even heterozygous mice were found to have a decreased survival rate after 2 weeks of gestation.[13] In humans, while deficiencies in CYPOR are not necessarily lethal, they do have some severe side effects, including disordered steroidogenesis and Antley-Bixler syndrome (ABS).[8][14] ABS is associated with urogenital defects (ie: ambiguous genitalia), cranial abnormalities (ie: brachycephaly) and skeletal defects (ie: bowed femurs, narrow ribcage and club feet), often due to disordered steroidogenesis.[8][14] Individuals with ABS and/or disordered steroidogenesis may have mutations in one or both alleles for CYPOR, although some cases are associated with mutations in another gene, fibroblast growth factor receptor 2 gene.[14][8]
References
- ↑ Horecker BL. Triphosphopyridine nucleotide-cytochrome c reductase in liver. J Biol Chem 1950 Apr 1;183(2):593-605
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 2.22 2.23 2.24 2.25 Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem. 2009 Apr 24;284(17):11374-84. Epub 2009 Jan 26. PMID:19171935 doi:10.1074/jbc.M807868200
- ↑ Phillips AH, Langdon RG. Hepatic triphosphopyridine nucleotide-cytochrome c reductase: Isolation, characterization, and kinetic studies. J Biol Chem 1962 Aug 1;237:2652-60
- ↑ 4.0 4.1 Smith GC, Tew DG, Wolf CR. Dissection of NADPH-cytochrome P450 oxidoreductase into distinct functional domains. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8710-4. PMID:8078947
- ↑ Li HC, Liu D, Waxman DJ. Transcriptional induction of hepatic NADPH: cytochrome P450 oxidoreductase by thyroid hormone. Mol Pharmacol. 2001 May;59(5):987-95. PMID:11306680
- ↑ Waxman DJ, Morrissey JJ, Leblanc GA. Hypophysectomy differentially alters P-450 protein levels and enzyme activities in rat liver: pituitary control of hepatic NADPH cytochrome P-450 reductase. Mol Pharmacol. 1989 Apr;35(4):519-25. PMID:2495435
- ↑ Ram PA, Waxman DJ. Thyroid hormone stimulation of NADPH P450 reductase expression in liver and extrahepatic tissues. Regulation by multiple mechanisms. J Biol Chem. 1992 Feb 15;267(5):3294-301. PMID:1737785
- ↑ 8.0 8.1 8.2 8.3 Tomkova M, Marohnic CC, Baxova A, Martasek P. [Antley-Bixler syndrome or POR deficiency?] Cas Lek Cesk. 2008;147(5):261-5. PMID:18630181
- ↑ 9.0 9.1 9.2 9.3 Nadler SG, Strobel HW. Identification and characterization of an NADPH-cytochrome P450 reductase derived peptide involved in binding to cytochrome P450. Arch Biochem Biophys. 1991 Nov 1;290(2):277-84. PMID:1929397
- ↑ 10.0 10.1 10.2 10.3 Tamburini PP, Schenkman JB. Differences in the mechanism of functional interaction between NADPH-cytochrome P-450 reductase and its redox partners. Mol Pharmacol. 1986 Aug;30(2):178-85. PMID:3016501
- ↑ Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995 Jan 15;3(1):41-62. PMID:7743131
- ↑ 12.0 12.1 Canessa M, Laski C, Falkner B. Red blood cell Na+ transport as a predictor of blood pressure response to Na+ load in young blacks and whites. Hypertension. 1990 Nov;16(5):508-14. PMID:2228151
- ↑ 13.0 13.1 13.2 Shen AL, O'Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2002 Feb 22;277(8):6536-41. Epub 2001 Dec 12. PMID:11742006 doi:10.1074/jbc.M111408200
- ↑ 14.0 14.1 14.2 Miller WL, Huang N, Pandey AV, Fluck CE, Agrawal V. P450 oxidoreductase deficiency: a new disorder of steroidogenesis. Ann N Y Acad Sci. 2005 Dec;1061:100-8. PMID:16467261 doi:1061/1/100
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |