Sandbox Reserved 198
From Proteopedia
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
Contents |
Semisynthetic Ribonuclease A
|
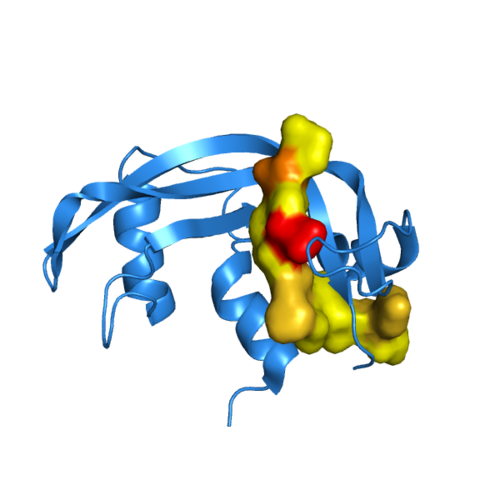
Above, is a two-dimesional representation of Semisynthetic RNase A. The synthetic peptide analog, RNase 111-118, is colored according to hydrophilicity. Yellow areas are comprised of hydrophobic residues. Red and brown segments are negatively and positively charged residues, respectively.
Introduction
The peptide synthesis of non-natural and non-coded proteins allowed scientists to analyze the mechanism and structure-activity relationships of classical enzyme molecules that were not accessible by traditional biomedical methods. These syntheses, though, were both difficult and time consuming, and advances in technique developed slowly[1]. At the beginning of the twentieth century, Emil Fischer performed the first synthesis of a peptide, but it was not until 1953 that the first peptide hormone was synthesized by Du Vigneaud[2]. The development of solid phase synthesis by Bruce Merrifield was a radical departure from traditional methods of bio-molecular synthesis that greatly increased efficiency. His method made possible the syntheses of much larger and more complex molecules; however, solid phase synthesis was not fully embraced until he demonstrated its full ability with the . This milestone synthesis and subsequent semisynthetic syntheses of enzymes including enriched the hypothesis that the amino acid sequence of a protein contains all necessary information to direct the formation of a fully active enzyme and, additionally, that an enzyme demonstrating the catalytic capacity and specificity of a naturally produced enzyme can be made in laboratory[3][4][5].
|
Structure equals Function
Semisynthetic RNase A
The synthesis of semisynthetic RNasa A clearly exhibits the structure to function relationship that defines proteins. In the RNase A protein, the removal of six C terminal residues, leaving , completely halts enzymatic activity[6]. However, a complex of RNase 1-118 with a synthetic polypeptide comprising the C terminal residues restores enzymatic activity to RNase A. Upon the addition of the synthetic chain, the adopts a structure that closely resembles that of [7]. The restoration of the structure reconstitutes the enzymatic activity of RNase to 98%[8].
Fully Synthetic RNase A
The demonstrates similar structural and functional characteristics (such as catalytic activity) as those of the RNase A[9]. The crystal data, X-ray data collection and refinement statistics show that the fully synthetic protein shares identical molecular structures with the wild type RNase A, and that the active sites of both emzymes contain no walter molecules and have no substrate ligand[10]. The crystal structure similarities of the , , and are further evidence that amino acid sequence dictates folded structure formation.
Synthetic Method
Solid-Phase Peptide Synthesis-Semisynthetic RNase A
Peptide synthesis is the production of proteins in which multiple amino acids are linked together through peptide bonds. A general chemical requirement for peptide synthesis is the blockage of the carboxyl group of one amino acid and the amino group of the second amino acid. The carboxyl group of the free carboxyl group can be activated and the new peptide bond is formed[11]. A common type of peptide synthesis is the solid-phase synthesis, in which the end of the peptide chain is attached to a solid support, as shown in Figure 1.
The semi-synthetic RNase A comprises of residues 1-118 and the synthetic analog of residues 111-124. The RNase 1-118 was prepared by successive digestion of RNase A pepsin and carboxypeptidase A[12]. The synthetic component, RNase 111-124, was prepared by the use of solid-phase peptide synthetic methods, in which the peptide chain was assembled in the stepwise manner while it was attached at one end to a solid support. The peptide chain was extended by repetitive steps of de-protection, neutralization and coupling until the desired sequence was obtained[13]. It was important that the synthesis proceeds rapidly and in high yields to prevent side reactions or by-products.
Peptide Ligation-Fully Synthetic RNase A
The peptide ligation chemistry in addition to solid-phase peptide synthesis is used to synthesize relatively longer peptide molecules with typical length of 125 residues[14]. The ligation methods overcome the length limitation of solid-phase synthesis, because the chemical ligation involves the joining of mutually reactive peptide segments created by solid-phase synthesis. The peptide bond in ligation is formed between an unprotected peptide and a peptide-thioester[15]. The shorter peptide segments are more rapidly prepared and are less susceptible to solubility issues in longer peptide chains.
The (124 residues) is prepared by two consecutive sets of one-pot ligations and related chemical transformations of six peptide segments (residues , , , , , , as highlighted in red)[16],which can prevent undesired byproduct formation. The six unprotected peptide segments were synthesized by highly optimized, stepwise solid-phase synthesis. This synthetic pathway is simple, has high overall yields, and it eliminate the need for the isolation of intermediate products.
Related Web-links
1. Introduction to Ribonuclease A by Raines: http://www.uta.edu/faculty/sawasthi/Enzymology-4351-5324/Class%20Syllabus%20Enzymology/ribonucleaseA.pdf 2. Introduction to Peptide Synthesis: http://en.wikipedia.org/wiki/Solid_phase_peptide_synthesis#Solid-phase_synthesis 3. Solid Phase Synthesis by Merrifield (Nobel Prize Winner):http://nobelprize.org/nobel_prizes/chemistry/laureates/1984/merrifield-lecture.pdf 4. Chemical Synthesis of Proteins:http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2845543/?tool=pmcentrez 5. Refined Crystal Structure: http://www.ncbi.nlm.nih.gov/pubmed/3680234
References
- ↑ Merrifield B. "Solid Phase Synthesis", Nobel Lecture, 8 December, 1984.
- ↑ Merrifield B. "Solid Phase Synthesis", Nobel Lecture, 8 December, 1984.
- ↑ Martin, Philip D., Marilynn S. Doscher, and Brian F. P. Edwards. "The Redefined Crystal Structure of a Fully Active Semisynthetic Ribonuclease at 1.8-A Resolution." The Journal of Biological Chemistry 262.33 (1987): 15930-5938.
- ↑ Merrifield B. "Solid Phase Synthesis", Nobel Lecture, 8 December, 1984.
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.
- ↑ Martin, Philip D., Marilynn S. Doscher, and Brian F. P. Edwards. "The Redefined Crystal Structure of a Fully Active Semisynthetic Ribonuclease at 1.8-A Resolution." The Journal of Biological Chemistry 262.33 (1987): 15930-5938.
- ↑ Martin, Philip D., Marilynn S. Doscher, and Brian F. P. Edwards. "The Redefined Crystal Structure of a Fully Active Semisynthetic Ribonuclease at 1.8-A Resolution." The Journal of Biological Chemistry 262.33 (1987): 15930-5938.
- ↑ Martin, Philip D., Marilynn S. Doscher, and Brian F. P. Edwards. "The Redefined Crystal Structure of a Fully Active Semisynthetic Ribonuclease at 1.8-A Resolution." The Journal of Biological Chemistry 262.33 (1987): 15930-5938.
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.
- ↑ Merrifield B. "Solid Phase Synthesis", Nobel Lecture, 8 December, 1984.
- ↑ Marilynn S. Doscher, Philip D. Martin and Brian F.P. Edwards, "Characerization of the Histidine Proton Nuclear Magnetic Resonance of a Semisynthetic Ribonuclease." Biochemistry, 1983,22,4125-4131
- ↑ Lin, M. C. (1970) Journal of Biological Chemistry, 245, 6726-6731
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.
- ↑ David J. Boerema, Valentina. A. T., Stephen B. H. Kent, "Total Synthesis by Modern chemical Ligation Methods and High Resolution (1.1-A) X-ray structure of Ribonuclease A. Biopolymers. 2008;90(3):278-86.