Sandbox Reserved 309
From Proteopedia
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
| |||||||||
| 2cdn, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Adenylate kinase, with EC number 2.7.4.3 | ||||||||
| Related: | 1p4s | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction/General Information
Adenylate kinase is a phosphotransferase that catalyzes the interconversion reaction of ATP, ADP, and AMP. Phosphotransferases include enzymes that catalyze phosphorylation reactions like the mechanism depicted below. It is part of the nucleotide and nucleoside kinases family. The adenylate kinase family can be classified into two groups: a long variant group, possessing a longer LID domain, and a short variant group where the Myobacterium tuberculosis bacterium can be found [1]. Since the source of this form of adenylate kinase is M. tuberculosis it is very important in developing treatment for tuberculosis in humans, as it is found to be essential for bacterial survival, and is therefore an excellent target for drugs when treating the disease. [1] Since tuberculosis is the leading cause of death from infectious disease worldwide, adenylate kinase is therefore a crucial protein in finding therapeutic tools required for prevention and treatment of this common and deadly disease. [2]
Structure
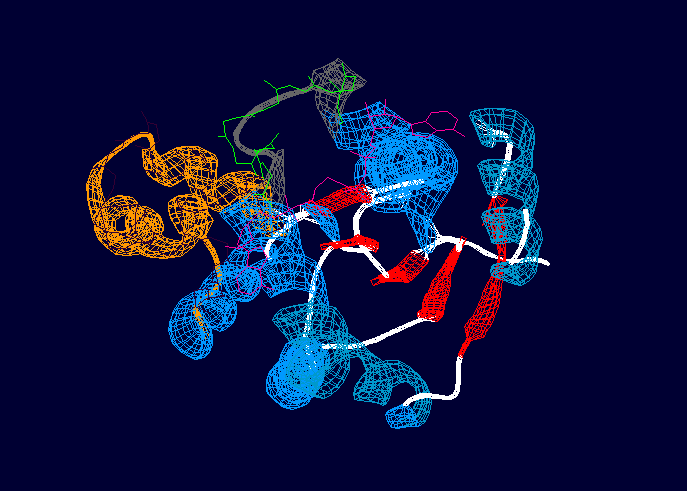
Protein of 201 residues with a consisting of two molecules of ADP (Adenosine-5'-Phosphate) and Magnesium ion.
|
The solution structure
With no ligand, adenylate kinase is represented by a central CORE domain composed of a 5-stranded parallel beta sheet surrounded by 7 alpha helices, and two periferal domains, LID and NMP [2], these two binding regions participate in the isolation of the substrates during catalysis and usually undergo significant conformational changes during the reaction [1]
The crystalline structure
Globular with a central core made by a surrounded by alpha helices, a P-loop motif at the N-terminus that binds ATP, and two regions (LID and NMP).[1] The LID region consists of a ten residue loop and is depicted in grey and green in the above image. The NMP-binding region is depicted in orange and dark purple. The overall rearrangement of the NMP binding region is 3.6 angstroms, and 6.4 angstroms in the LID region, indicating that these two regions experience large conformational changes during activation. [1]
Function
- Involved in nucleotide biosynthesis, which is very important as the intervention point for therapeutic agents.
- Catalyzes the reversible Mg2+ dependent transfer of the terminal phosphate group from ATP to AMP releasing two molecules of ADP or vice versa.
- During the reaction, a molecule of ADP binds to the ATP-binding site, while the NMP-binding site binds either ADP or AMP. The ADP molecule is then phosphorylated in the ATP site and AMP is formed in the NMP site. [1]
- LID and NMP binding regions go through a significant conformational change during the catalysis reaction. [1]
Catalytic Mechanism
The catalytic mechanism facilitated by adenylate kinase is depicted below:
ADP+MgADP↔MgATP+AMP
The geometry of the surrounding amino acids, and the distribution of positive charges of the active sites suggest that there is a direct nucleophilic attack by the oxygen on the donor substrate. Together with the Mg2+ ion, the surrounding side chains are positioned to neutralize the negative transition state and allow the positive polarization required to transfer the phoshporus atom from one of the ADP molecules and therefore create ATP and AMP. [1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Bellinzoni M, Haouz A, Grana M, Munier-Lehmann H, Shepard W, Alzari PM. The crystal structure of Mycobacterium tuberculosis adenylate kinase in complex with two molecules of ADP and Mg2+ supports an associative mechanism for phosphoryl transfer. Protein Sci. 2006 Jun;15(6):1489-93. Epub 2006 May 2. PMID:16672241 doi:10.1110/ps.062163406
- ↑ 2.0 2.1 Miron S, Munier-Lehmann H, Craescu CT. Structural and dynamic studies on ligand-free adenylate kinase from Mycobacterium tuberculosis revealed a closed conformation that can be related to the reduced catalytic activity. Biochemistry. 2004 Jan 13;43(1):67-77. PMID:14705932 doi:10.1021/bi0355995