Sandbox Reserved 315
From Proteopedia
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
Introduction
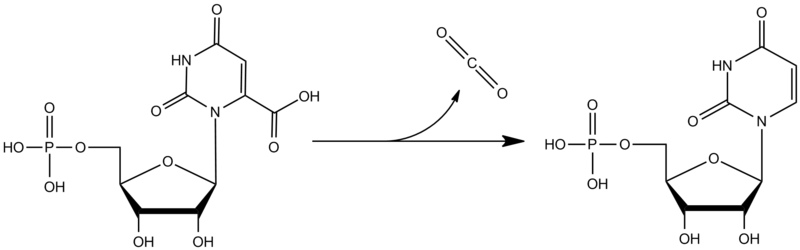
Orotidine monophosphate decarboxylase (ODCase), also known as orotidine 5’-monophosphate decarboxylase (OMP decarboxylase) or orotidine 5’-phosphate decarboxylase, is an enzyme involved in nucleic acid biosynthesis. It catalyzes the conversion of orotidine 5’-monophosphate (OMP) to uridine 5’-monophosphate (UMP). This reaction is the final step in de novo pyrimidine nucleotide synthesis and is an essential precursor for both DNA and RNA. The main structure of orotidine monophosphate decarboxylase is a TIM barrel, with one side acting as the binding site, while the other side is closed-off. ODCase is often bound to other proteins in biological conditions. In single-cellular organisms, it can form a dimeric enzyme by binding to another ODCase[1]. In multi-cellular organisms it can bind to orotate phosphoribosyltransferase to form a bifunctional protein[2]. Additionally this enzyme has been studied for it’s remarkable catalytic efficiency[3].
Contents |
Structure
|
| |||||||||
| 1dv7, resolution 1.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Activity: | Orotidine-5'-phosphate decarboxylase, with EC number 4.1.1.23 | ||||||||
| Related: | 1dvj | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Orotidine monophosphate decarboxylase has a TIM barrel structure[1][3]. Like a typical TIM barrel, it is cylindrically-shaped as a result of parallel α helices and β sheets arranged in a circular manner. In biological conditions, ODCase is found in dimeric form, covalently bonded to a second ODCase[3]. Each ODCase has 9 α helices that encompass 8 internal β sheets[1][3]. Both the C and N terminus are oriented on the same side of the monomer and directed away from the interface between the two monomers; this could explain how the enzymes can still maintain functionality when bound to another protein[3]. The loops connecting these α helices and β sheets are where the active sites are found[3]. The active site is only found on one side of the barrel, the “open” side, while the other side is closed off[3].
Active Site
The is located in the opening of the TIM barrel. The ligand binds through extensive electrostatic interactions and hydrogen bonding with the ribose ring and phosphate group of OMP[1]. Additionally, the active site forms a hydrophobic region around the pyrimidine ring of OMP [1]. A unique feature of the active site is the series of alternating lysine and asparatate groups[1]. This combination of basic and acidic groups contributes to binding, catalysis, and product release[1]. The active site has both a hydrophobic pocket, which surrounds the pyrimidine base, and a hydration shell, which interacts with the phosphate group[1]. This hydration shell involves several water molecules acting as bridges between the substrate and active site.
Function
Orotidine monophosphate decarboxylase is one of several proteins in de novo pyrimidine biosynthesis. Orotidine monophosphate (OMP) is formed when orotate reacts with 5-phosphoribosyl α-diphosphate (PRPP)[4]. ODCase then decarboxylated OMP to form uridine monophosphate (UMP), which then goes on to be phosphorylated and converted into cytosine, uracil, and thymine[1][4]. The mechanism for decarboxylation of OMP is still unclear. Unlike other biochemical decarboxylation reactions that stabilize the reaction intermediate through delocalizing the electron pair, OCDase shows no such stabilization[5]. One possible mechanism is the formation of a stabilized carbene intermediate resulting from protonization at the 4-C position of OMP[5].
Biological Significance
All organisms carry out pyrimidine biosynthesis and therefore require orotidine monophosphate decarboxylase. ODCase activity is competitively inhibited by UMP and so partly regulates pyrimidine synthesis. Mutations in ODCase are genetically inherited and can cause orotic aciduria[5][4]. This disorder is identified by orotic acid accumulation in the urine and can cause physical retardation and anemia[4].
UMP Synthase
In multicellular eukaryotes, orotidine monophosphate decarboxylase associates with orotate phosphoribosyltransferase to form a bifunctional protein, uridine monophosphate synthase uridine monophosphate synthase (UMP synthase)[5][2]. UMP synthase carries out the last two steps in pyrimidine biosynthesis, converting orotate to uridine 5'-monophosphate[2]. This reaction involves first adding ribose-P to orotate to form orotidine 5'-monophosphate, followed by a decarboxylation reaction to form uridine 5'-monophosphate[2]. In microorganisms these two enzymes are separate and coded by distinct genes. However research has shown that all known multicellular eukaryotes code the genes for these two enzymes together and as a result they are covalently bonded as a bifunctional protein with two distinct catalytic domains[2]. UMP synthase likely evolved in multicellular organisms for several reasons. It may increase efficiency of nucleotide synthesis by channeling the OMP intermediate from the first enzyme directly to the next[2]. Similarly, it improves allosteric control between the two domains[2]. It has also been shown that having a dimeric enzyme improves stability of both domains in UMP synthase compared to separate enzymes[2].
Rate of Catalysis
ODCase is a strong catalyst for OMP decarboxylation. The decarboxylation of OMP to UMP has a halftime of 18 milliseconds when catalyzed by ODCase[3]. For this reaction to occur spontaneously it would take an astounding halftime of 78 million years[3]. This equates to a catalytic proficiency of 2.0 x 1023 M-1, making ODCase one of the most proficient catalytic enzymes known[5]. Additionally, ODCase achieves this catalytic power without the use of metals or other cofactors[1][3]. Instead, the catalytic efficiency is mainly achieved through destabilizing the reactive group on the substrate[1].
Substrate Destabilization
OMP is strongly attracted to the active site and further stabilized by phosphate and ribose interactions with ODCase[1]. However, the C¬¬6 carboxylate anion is strongly destabilized through repulsion by Asp70 and it is this destabilization that favors the decarboxylation reaction[1]. The simultaneous attraction and repulsion between the enzyme and substrate is known as the “Circe effect”, as described by William Jencks[1].
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Wu N, Mo Y, Gao J, Pai EF. Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2017-22. PMID:10681441 doi:10.1073/pnas.050417797
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Yablonski MJ, Pasek DA, Han BD, Jones ME, Traut TW. Intrinsic activity and stability of bifunctional human UMP synthase and its two separate catalytic domains, orotate phosphoribosyltransferase and orotidine-5'-phosphate decarboxylase. J Biol Chem. 1996 May 3;271(18):10704-8. PMID:8631878
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Anatomy of a proficient enzyme: the structure of orotidine 5'-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2011-6. PMID:10681417 doi:10.1073/pnas.030409797
- ↑ 4.0 4.1 4.2 4.3 Brosnan ME, Brosnan JT. Orotic acid excretion and arginine metabolism. J Nutr. 2007 Jun;137(6 Suppl 2):1656S-1661S. PMID:17513443
- ↑ 5.0 5.1 5.2 5.3 5.4 Lee JK, Houk KN. A proficient enzyme revisited: the predicted mechanism for orotidine monophosphate decarboxylase. Science. 1997 May 9;276(5314):942-5. PMID:9139656