Sandbox Reserved 327
From Proteopedia
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
Yeast eIF1
Contents |
Introduction
Translation is an essential process for both prokaryotes and eukaryotes to make various proteins from nucleic acid. It requires three different steps: initiation, elongation, and termination. Many proteins are needed in the initiation phase to form an initiation complex with the ribosome and promote the translation. In eukaryotes, these proteins are also known as eukaryotic initiation factors (eIF). The eIFs were first studied through the genetic analyzed of yeast Saccharomyces cerevisiae showed that the initiation process is complex since it involves at least 12 eIFs containing more than 30 polypeptides , including eIF1, eIF2, eIF3, eIF4, and eIF5 [1].
Structure and the binding sites
| |||||||||
| 2ogh, 20 NMR models () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | SUI1, RFR1 (Saccharomyces cerevisiae) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
eIF1 is a small protein (12 kDa) that is encode by sui1 which is one of the component of multifactor complex (MFC) that plays an important role in regulating translation initiation [2][3]. eIF1 is a universal translation factor across organisms which make eIF1 homologs can be found in other eukayotes, archaea, and some bacteria [4]. Yeast eIF1 contains a on one side with N-terminal tail (aa 1-23) or NTT [2]. It is homolog to since it has 87% structure similarity and 63% identity (DNA sequence) to the human eIF1 [2] [4]. However, yeast eIF1 has two different conformations with two clear sets of backbone resonances that interconverting with each other and 20 different possible models for the N-terminal tail structure [2]. So far, only the binding sites of eIF1 to ribosome and eIF5 have been determined, while the exact binding site interactions with other initiation proteins are still unknown [2] [4].
eIF1-ribosome binding site
eIF1 would bind near the P-site region of the ribosomal 40S, changes the conformation of the 40S subunit, the position of mRNA and the start codon initiation [5]. This binding site region is unexpectedly similar to the binding site of prokaryotic initiation factor IF3 to the ribosome, despite their unrelated structure [5]. Together with eIF1A, eIF5, eIF3, and the eIF2.GTP.Met-tRNAiMet ternary complex (TC), eIF1 would form a 43S preinitiation complex (PIC) or also known as multi factor complex (MFC)[2][5][6].
eIF1-eIF5 binding sites
There are two : at the NTT site of eIF1 and at a KH surface area, a specific region that rich in Lysine (K) and hydrophobic (H) residues [2]. These bindings are different from the eIF1-ribosome binding site and are salt dependent, with high salt concentration would make the interaction weaker [2].
eIF1-NTT
The eIF1-NTT area is believed to play role in stimulating the MFC assembly by promoting the eIF5- eIF2β [2]. In vitro mutation in the eIF1-NTT area would alter the binding of eIF5 and eIF2β, but not eIF3c which supported that this area played role in stimulating the MFC assembly [2]. In vivo study also supported that eIF1-NTT would allow the binding of eIF1 to the eIF5 and eIF2β [2].
eIF1-KH
The other binding site area of eIF1 to eIF5 which is believed to play role in the AUG start codon selection since in vitro mutation in this area by altering the basic part of the KH region would relax the start codon selection [2].The mutation would also alter the formation of MFC, but not like mutation in the eIF1-NTT area, eIF1-NTT mutation is lethal to the yeast growth [2]. Mutation that alter the hydrophobic residue would disrupt a critical link to the PIC and prevent the eIF1 dissociation [6]. The critical link is believed to be strongly associate with eIF5 [2].
Function and Mechanism
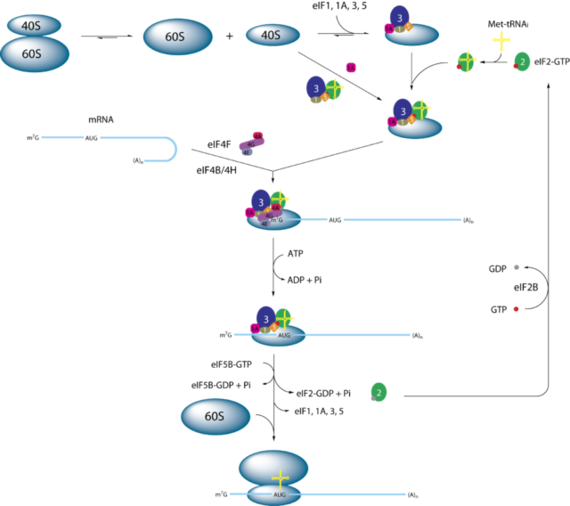
eIF1 is essential to control the ribosome conformational rearrangement in translation initiation by stimulating the formation of MFC and controlling the start codon selection which allow the translation elongation process to occur [2][3].Start codon selection is a key step for translation elongation to occur because recognition of the right AUG start codon would lead to the binding of 60S subunit to form the 80 S initiation complex, a precursor for elongation [2]. Study showed eIF1 dissociation plays a critical role in start codon selection [6].
Initially, eIF1 in the 43S PIC would interact with the 5’end of mRNA to form 48S PIC and promotes scanning until the Met-tRNAiMet recognizes the right AUG codon from the mRNA [2][6]. Once the right codon is recognized, there would be a conformational change on the ribosome and the Met-tRNAiMet would be released to the P-site, ready for the elongation process (paper, cheung). Also, hydrolysis of eIF2.GTP to GDP by the action of N-terminal residues of eIF5 in the PIC complex would occur, leading to a Pi being released [2][6]. These two events would let the eIF1 and eIF2.GDP to be released from the PIC. Then, eIF5 GTPase would promote the binding of the ribosomal 60S forming the 80S ribosome, get it ready for elongation [2]. The eIF2-GDP is then going to be re-used for another TC formation when it is recycled back to eIF2-GTP by eIF2B.GEF [2]. However, when the codon-anticodon pair is mismatched, eIF1 would avoid the recognition by preventing the hydrolysis through blocking the Pi release at non-AUG codons and repressing the activity of eIF5 GTPase [2][5][6].
Mutations in eIF1 would alter the function and mechanism of eIF1 in the cell. There are two classesof mutations in yeast eIF1: Mof2 and Sui1 mutations [4]. Mof2 mutation in eIF1 would increase the ribosome frameshifting in the translation process [4]. As for Sui1 mutation, in vitro and in vivo mutations would let the mismatched pairing of Met-tRNAiMet with non-AUG codons to occur [4]. Initiation at UUG codons would be increased instead, while the binding of eIF1 to 40S subunits would be reduced [6]. However, this mutation would increase the eIF1 dissociation and Pi release rate from the PIC while another mutation that is completely the opposite, suppress the expression of Sui mutation (hyperaccuracy mutation in eIF1A), would decrease the dissociation rate [6].
See also
References
- ↑ 1.0 1.1 Maduzia LL, Moreau A, Poullet N, Chaffre S, Zhang Y. The role of eIF1 in translation initiation codon selection in Caenorhabditis elegans. Genetics. 2010 Dec;186(4):1187-96. Epub 2010 Sep 20. PMID:20855569 doi:10.1534/genetics.110.121541
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem. 2008 Jan 11;283(2):1094-103. Epub 2007 Nov 1. PMID:17974565 doi:10.1074/jbc.M708155200
- ↑ 3.0 3.1 3.2 Asano K, Sachs MS. Translation factor control of ribosome conformation during start codon selection. Genes Dev. 2007 Jun 1;21(11):1280-7. PMID:17545463 doi:10.1101/gad.1562707
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999 May 4;18(9):2631-7. PMID:10228174 doi:10.1093/emboj/18.9.2631
- ↑ 5.0 5.1 5.2 5.3 5.4 Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003 Nov 15;17(22):2786-97. Epub 2003 Nov 4. PMID:14600024 doi:10.1101/gad.1141803
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007 May 15;21(10):1217-30. PMID:17504939 doi:10.1101/gad.1528307

