Context
The SUMO-conjugating enzyme UBC9 is involved in ubiquitination of proteins.
The Ubiquitin is a small protein of 76 aa.
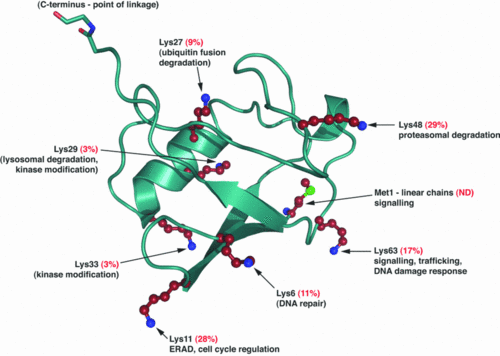
Ubiquitination is a post translationnal modification where an ubiquitin is attached to a protein. This modification has several consequences, it can lead to the degradation of the protein via the proteasome, it can change the protein localization or it can alter the interaction of the protein with other factors, this is why this modification plays an important role in the control of many cellular processes.
UBC9, in particular, is important in the cell cycle progression because it allows the degradation of S phase and M phase cyclines.
Ubiquitination occurs in three steps : Activation performed by ubiquitin-activating enzymes E1, conjugation performed by ubiquitin-conjugating enzymes E2 and ligation performed by ubiquitin ligases E3.
During the first step the Ct Carboxyl group of the ubiquitin is linked to the cystein sulfhydryl group of E1.
The second step is a transesterification to transfer the active ubiquitin from the cystein of E1 to the cystein of E2. E2 binds the activated ubiquitin and E1.
During the third step E3 catalyses the formation of an isopeptide bond between a lysine of the substrate and a glycine of the ubiquitin.
UBC9 is a E2. It is a lynchpin in the SUMO pathway, it interacts with E1 during the activation, then with the ubiquitin after the transfer and finally with E3 during conjugation. It is particularly important for the formation of polymeric chain when the number of SUMO exceeds 2.
Structure
UBC9 is a monomer of 158 amino acids.
It is comprised of a single domain with and , typical of the core domain of the UBCs.
The dimensions of the surface are 20 Å X 30 Å X 50 Å.
The core domain contains an (β1 to β4, residues 25-30, 36-46, 57-63, 74-77) and (α1 to α4, residues 1-18, 109–121, 131-139, 141-154)
This secondary structure represent 50% of the polypetide (33% α-helices and 17% β-sheets)
There are : Pro69 and Pro79 that play an important role in the structure of the protein.
Function
The active site residue is located in a formed by a loop between β4 and α2 (85-102) and another one between α2 and α3 (residues 122-130).
are the most likely to participate to the catalytic action because they are located within 6 Å of the sulfhydryl group of the accepting cysteine and that their side chains are oriented toward the Cysteine : Asn85, Tyr87, Glu98, Lys101, and Asp127.
are well conserved among the UBC family : Gly47, Lys48, Gly56, Tyr68, Pro69, Pro73, Phe77, His83, Pro84, Asn85, Gly90, Trp103, Pro105, Leu120 and Pro128.
Most of them are non polar and they are unlikely to have a direct role in the catalytic action but they are probably positionned to maintain the special configuration of the active site.
Interaction with the substrates
Two insertions have been observed by comparing UBC9 with other UBCs :
- 5 residues (32-36) form most of a very exposed that connects strands β1 and β2.
- The residues Asp100 and Lys101 form a (residues 94–102) close to Cys93
Those insertions provide additionnal binding sites for new substrate without blocking the access to Cys
The Surface electric potential :
There is a negative patch surrounding the active site that is conserved between all UBCs, it is probably important in the interaction with Ubiquitin and E1, the common substrates of all UBC9s.
But there are many particularities in the surface electric potential of UBC9, that probably reflect the specificity for E3 and the protein substrate. The electrostatic dipole is more important for UBC9 than for others UBCs. The positive charged are located on the back face of the molecule. One, for example, is located on the N-terminal region, it is composed of a segment of basic residues separated by nonpolar residues (13RKAWRK18). This , the presence of those two specifities in the same region could be a sign that this site is responsible for the specificity of UBC9 for some particular E3 and protein substrates.

