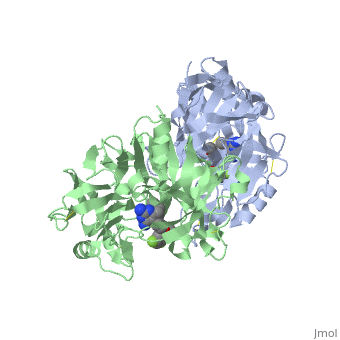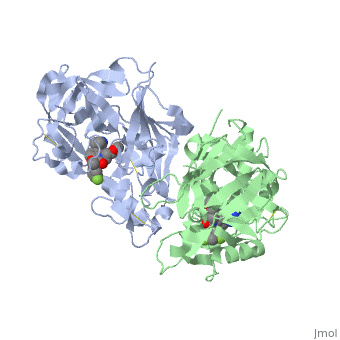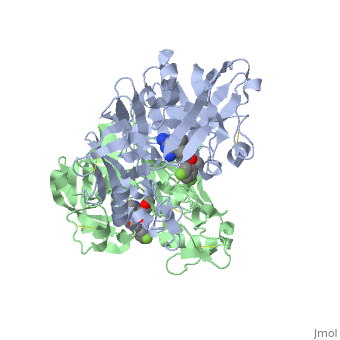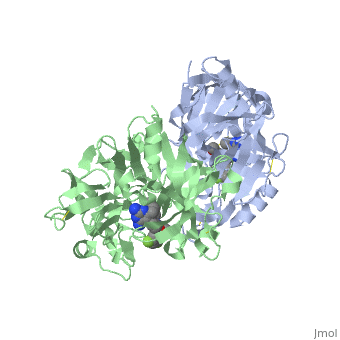We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
CHEM2052 Tutorial Example4
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 25: | Line 25: | ||
This view shows the amino acid residues on the <scene name='59/596437/Renin_interactions_rhs/2'>right hand side</scene> of your "map". You should notice that these amino acids are fairly hydrophobic and interact with the hydrophobic parts of the inhibitor. If you prefer to view the structure where the inhibitor atoms are colour coded you will <scene name='59/596437/Renin_interactions_rhs_again/1'>find it here. </scene> | This view shows the amino acid residues on the <scene name='59/596437/Renin_interactions_rhs/2'>right hand side</scene> of your "map". You should notice that these amino acids are fairly hydrophobic and interact with the hydrophobic parts of the inhibitor. If you prefer to view the structure where the inhibitor atoms are colour coded you will <scene name='59/596437/Renin_interactions_rhs_again/1'>find it here. </scene> | ||
| - | Similarly this view shows the amino acid residues on the | + | Similarly this view shows the amino acid residues on the |
| + | <scene name='59/596437/Lhs_aminoacids_representation/2'>left hand side</scene> of your "map". Again you can view this with the atoms of the inhibitor colour coded | ||
| + | <scene name='59/596437/Lhs_aminoacids_representation/3'>find it here</scene>. | ||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | |||
| - | |||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
CHEM2052_Tutorial_Example4
| |||||||||||