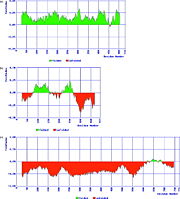
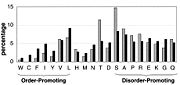
Intrinsically Disordered Protein
From Proteopedia
(Redirected from Intrinsically Unfolded Proteins (IUP))
| |||||||||||
References and Notes
- ↑ For the sake of brevity, this description is oversimplified. RNAse needed to be reduced to break disulfide bonds, as well as using 8 M urea, for denaturation. Oxidation without the denaturant then left an inactive enzyme because the disulfide bonds formed randomly, precluding proper folding except very slowly (many hours). Only when protein disulfide isomerase was added did the re-folding occur at a physiological rate (about a minute). The fact that RNAse could thus be trapped in an inactive conformation under physiological conditions contributed to the insights developed by Anfinsen and his team. Proteins lacking disulfides renatured in seconds. For details, see Anfinsen's Nobel Lecture.
- ↑ A similar observation was made around the same time by then graduate student Lisa Steiner in the lab of Fred Richards at Yale University. Neither Richards nor advisor Joseph Fruton thought the observation interesting enough to publish. It was an answer to a question not yet asked. This story is recounted by David Eisenberg, see the next citation.
- ↑ Eisenberg DS. How Hard It Is Seeing What Is in Front of Your Eyes. Cell. 2018 Jun 28;174(1):8-11. doi: 10.1016/j.cell.2018.06.027. PMID:29958112 doi:http://dx.doi.org/10.1016/j.cell.2018.06.027
- ↑ Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999 Oct 22;293(2):321-31. PMID:10550212 doi:10.1006/jmbi.1999.3110
- ↑ Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26-59. PMID:11381529
- ↑ Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002 Jan;269(1):2-12. PMID:11784292
- ↑ 7.0 7.1 7.2 7.3 Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002 Oct;27(10):527-33. PMID:12368089
- ↑ Summary of the previous paper (Tompa, 2002): The disorder of intrinsically disordered proteins (IDP's) is crucial to their functions. They may adopt defined but extended structures when bound to cognate ligands. Their amino acid compositions are less hydrophobic than those of soluble proteins. They lack hydrophobic cores, and hence do not become insoluble when heated. About 40% of eukaryotic proteins have at least one long (>50 residues) disordered region. Roughly 10% of proteins in various genomes have been predicted to be fully disordered. Presently over 100 IDP's have been identified; none are enzymes. Obviously, IDP's are greatly underrepresented in the Protein Data Bank, although there are a few cases of an IDP bound to a folded (intrinsically structured) protein. Here, Tompa suggests five functional categories for intrinsically unstructured proteins and domains: entropic chains (bristles to ensure spacing, springs, flexible spacers/linkers), effectors (inhibitors and disassemblers), scavengers, assemblers, and display sites. (Summary by Eric Martz.)
- ↑ Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008 Dec;18(6):756-64. Epub 2008 Nov 17. PMID:18952168 doi:10.1016/j.sbi.2008.10.002
- ↑ Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004 Aug;18(11):1169-75. PMID:15284216 doi:10.1096/fj.04-1584rev
- ↑ Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci. 2003 Feb;28(2):81-5. PMID:12575995
- ↑ Summary of the previous paper (Gunasekaran et al., 2003): Argues that proteins involved in extensive protein-protein interactions can function effectively despite having their structure depend upon such interactions, so that as monomers they are natively disordered. Dispensing with the structural framework (scaffold) needed to maintain a stable fold in the monomer increases efficiency by reducing size. This may account for the large percentage (roughly half) of all proteins that are predicted to be natively disordered. (Summary by Eric Martz.)
- ↑ Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005 Mar;6(3):197-208. PMID:15738986 doi:10.1038/nrm1589
- ↑ 14.0 14.1 14.2 Oldfield CJ, Xue B, Van YY, Ulrich EL, Markley JL, Dunker AK, Uversky VN. Utilization of protein intrinsic disorder knowledge in structural proteomics. Biochim Biophys Acta. 2013 Feb;1834(2):487-98. doi: 10.1016/j.bbapap.2012.12.003., Epub 2012 Dec 8. PMID:23232152 doi:http://dx.doi.org/10.1016/j.bbapap.2012.12.003
- ↑ 15.0 15.1 Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575-8. PMID:2145515 doi:http://dx.doi.org/10.1038/347575a0
- ↑ Pontius BW, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8403-7. PMID:2236048
- ↑ For the unstructured domain interpretation of early work by Pontius and Berg, see the 2004 review by Tompa and Csermley, PMID: 15284216
- ↑ Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008 Sep 16;9 Suppl 2:S1. PMID:18831774 doi:10.1186/1471-2164-9-S2-S1
- ↑ Martz, E. Book review of Introduction to protein science—architecture, function, and genomics: Lesk, Arthur M.. Biochem. Mol. Biol. Educ. 33:144-5 (2006). DOI: 10.1002/bmb.2005.494033022442
- ↑ Chouard T. Structural biology: Breaking the protein rules. Nature. 2011 Mar 10;471(7337):151-3. PMID:21390105 doi:10.1038/471151a
- ↑ Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161-71. PMID:11700597
- ↑ Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11504-9. PMID:8876165
- ↑ Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. J Mol Biol. 2002 Oct 4;322(5):917-27. PMID:12367518
- ↑ Schweers O, Schonbrunn-Hanebeck E, Marx A, Mandelkow E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J Biol Chem. 1994 Sep 30;269(39):24290-7. PMID:7929085
- ↑ Tompa P, Kovacs D. Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol. 2010 Apr;88(2):167-74. doi: 10.1139/o09-163. PMID:20453919 doi:http://dx.doi.org/10.1139/o09-163
- ↑ Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999 Feb;6(2):117-23. PMID:10048921 doi:10.1038/5803
- ↑ Markus MA, Hinck AP, Huang S, Draper DE, Torchia DA. High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA. Nat Struct Biol. 1997 Jan;4(1):70-7. PMID:8989327
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
- ↑ Schmitz ML, dos Santos Silva MA, Altmann H, Czisch M, Holak TA, Baeuerle PA. Structural and functional analysis of the NF-kappa B p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J Biol Chem. 1994 Oct 14;269(41):25613-20. PMID:7929265
- ↑ Baskakov IV, Kumar R, Srinivasan G, Ji YS, Bolen DW, Thompson EB. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem. 1999 Apr 16;274(16):10693-6. PMID:10196139
- ↑ Bondos SE, Swint-Kruse L, Matthews KS. Flexibility and Disorder in Gene Regulation: LacI/GalR and Hox Proteins. J Biol Chem. 2015 Oct 9;290(41):24669-77. doi: 10.1074/jbc.R115.685032. Epub 2015, Sep 4. PMID:26342073 doi:http://dx.doi.org/10.1074/jbc.R115.685032
- ↑ Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13452-7. PMID:9391046
- ↑ Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT Jr. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996 Oct 29;35(43):13709-15. PMID:8901511 doi:10.1021/bi961799n
- ↑ Dunker AK. Another disordered chameleon: the Micro-Exon Gene 14 protein from Schistosomiasis. Biophys J. 2013 Jun 4;104(11):2326-8. doi: 10.1016/j.bpj.2013.04.018. PMID:23746503 doi:http://dx.doi.org/10.1016/j.bpj.2013.04.018
- ↑ 35.0 35.1 35.2 35.3 35.4 35.5 Tsuboyama K, Osaki T, Matsuura-Suzuki E, Kozuka-Hata H, Okada Y, Oyama M, Ikeuchi Y, Iwasaki S, Tomari Y. A widespread family of heat-resistant obscure (Hero) proteins protect against protein instability and aggregation. PLoS Biol. 2020 Mar 12;18(3):e3000632. doi: 10.1371/journal.pbio.3000632., eCollection 2020 Mar. PMID:32163402 doi:http://dx.doi.org/10.1371/journal.pbio.3000632
- ↑ Isoelectric points were estimated with the Protein Calculator.
- ↑ 37.0 37.1 Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005 Aug 15;21(16):3435-8. Epub 2005 Jun 14. PMID:15955783 doi:http://dx.doi.org/10.1093/bioinformatics/bti537
- ↑ Zeev-Ben-Mordehai T, Rydberg EH, Solomon A, Toker L, Auld VJ, Silman I, Botti S, Sussman JL. The intracellular domain of the Drosophila cholinesterase-like neural adhesion protein, gliotactin, is natively unfolded. Proteins. 2003 Nov 15;53(3):758-67. PMID:14579366 doi:10.1002/prot.10471
- ↑ 39.0 39.1 Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001 Sep;19(9):805-6. PMID:11533628 doi:10.1038/nbt0901-805
- ↑ Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001 Jan 1;42(1):38-48. PMID:11093259
- ↑ Noivirt-Brik O, Prilusky J, Sussman JL. Assessment of disorder predictions in CASP8. Proteins. 2009 Aug 21. PMID:19774619 doi:10.1002/prot.22586
- ↑ Hu G, Katuwawala A, Wang K, Wu Z, Ghadermarzi S, Gao J, Kurgan L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat Commun. 2021 Jul 21;12(1):4438. PMID:34290238 doi:10.1038/s41467-021-24773-7
- ↑ Necci M, Piovesan D, Tosatto SCE. Critical assessment of protein intrinsic disorder prediction. Nat Methods. 2021 May;18(5):472-481. PMID:33875885 doi:10.1038/s41592-021-01117-3
- ↑ Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005 Aug 15;21(16):3369-76. Epub 2005 Jun 9. PMID:15947016 doi:http://dx.doi.org/10.1093/bioinformatics/bti534
- ↑ Holladay NB, Kinch LN, Grishin NV. Optimization of linear disorder predictors yields tight association between crystallographic disorder and hydrophobicity. Protein Sci. 2007 Oct;16(10):2140-52. PMID:17893360 doi:16/10/2140
- ↑ Denning DP, Uversky V, Patel SS, Fink AL, Rexach M. The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J Biol Chem. 2002 Sep 6;277(36):33447-55. Epub 2002 Jun 13. PMID:12065587 doi:10.1074/jbc.M203499200
- ↑ Xue B, Brown CJ, Dunker AK, Uversky VN. Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim Biophys Acta. 2013 Apr;1834(4):725-38. doi: 10.1016/j.bbapap.2013.01.012., Epub 2013 Jan 22. PMID:23352836 doi:http://dx.doi.org/10.1016/j.bbapap.2013.01.012
- ↑ 48.0 48.1 48.2 Hsu WL, Oldfield CJ, Xue B, Meng J, Huang F, Romero P, Uversky VN, Dunker AK. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013 Mar;22(3):258-73. doi: 10.1002/pro.2207. Epub 2013 Jan 27. PMID:23233352 doi:http://dx.doi.org/10.1002/pro.2207
- ↑ Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996 Jul 25;382(6589):325-31. PMID:8684460 doi:10.1038/382325a0
- ↑ Konig P, Richmond TJ. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139-54. PMID:8377181 doi:http://dx.doi.org/10.1006/jmbi.1993.1490
- ↑ Hollenbeck JJ, McClain DL, Oakley MG. The role of helix stabilizing residues in GCN4 basic region folding and DNA binding. Protein Sci. 2002 Nov;11(11):2740-7. PMID:12381856 doi:http://dx.doi.org/10.1110/ps.0211102
See Also
- Temperature value
- Temperature color schemes
- Temperature value vs. resolution
- Resolution
- Quality assessment for molecular models
- NMR Ensembles of Models
- Anisotropic refinement
Authorship
The bulk of this article was written by Tzviya Zeev-Ben-Mordehai. Contributions by Eric Martz were minor -- his name is listed first due to a technicality.
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Wayne Decatur, Alexander Berchansky, Joel L. Sussman, Tzviya Zeev-Ben-Mordehai, Michal Harel, David Canner, Karl Oberholser, Jaime Prilusky, Eran Hodis