Structural highlights
Function
NADM_METTH
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
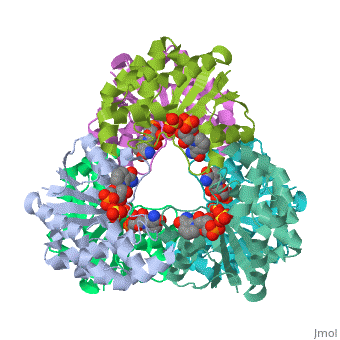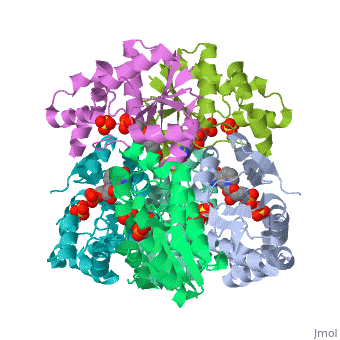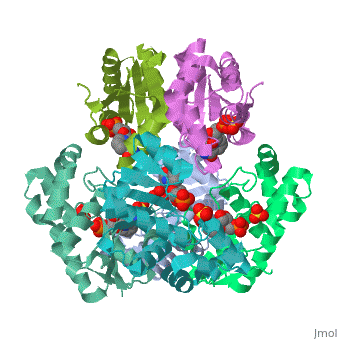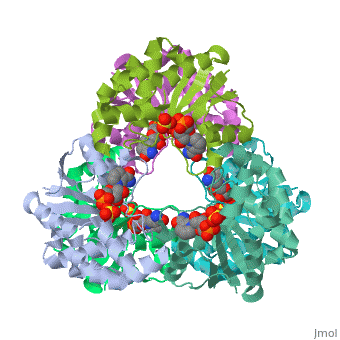
Nicotinamide mononucleotide adenylyltransferase (NMNATase) catalyzes the linking of NMN(+) or NaMN(+) with ATP, which in all organisms is one of the common step in the synthesis of the ubiquitous coenzyme NAD(+), via both de novo and salvage biosynthetic pathways. The structure of Methanobacterium thermoautotrophicum NMNATase determined using multiwavelength anomalous dispersion phasing revealed a nucleotide-binding fold common to nucleotidyltransferase proteins. An NAD(+) molecule and a sulfate ion were bound in the active site allowing the identification of residues involved in product binding. In addition, the role of the conserved (16)HXGH(19) active site motif in catalysis was probed by mutagenic, enzymatic and crystallographic techniques, including the characterization of an NMN(+)/SO4(2-) complex of mutant H19A NMNATase.
Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.,Saridakis V, Christendat D, Kimber MS, Dharamsi A, Edwards AM, Pai EF J Biol Chem. 2001 Mar 9;276(10):7225-32. Epub 2000 Nov 3. PMID:11063748[1]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Saridakis V, Christendat D, Kimber MS, Dharamsi A, Edwards AM, Pai EF. Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes. J Biol Chem. 2001 Mar 9;276(10):7225-32. Epub 2000 Nov 3. PMID:11063748 doi:10.1074/jbc.M008810200