5d9y
From Proteopedia
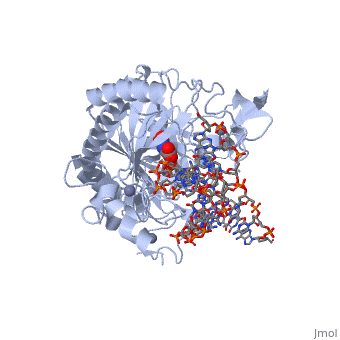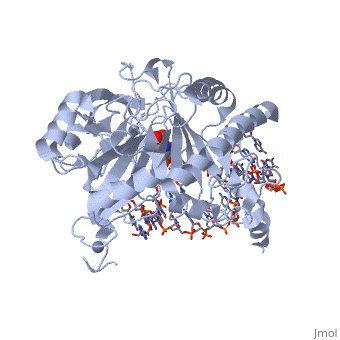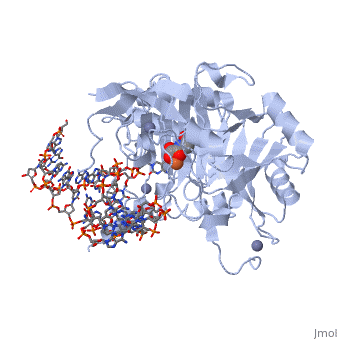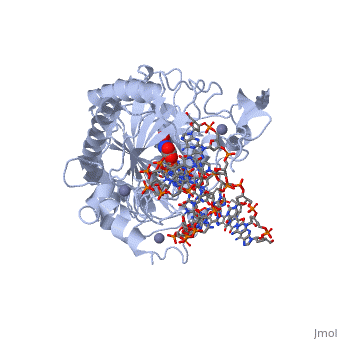
Crystal structure of TET2-5fC complex
Structural highlights
DiseaseTET2_HUMAN Refractory anemia;Polycythemia vera;Acute myeloid leukemia with multilineage dysplasia;Essential thrombocythemia;Myelofibrosis with myeloid metaplasia;Refractory anemia with excess blasts;Acquired idiopathic sideroblastic anemia. TET2 is frequently mutated in myeloproliferative disorders (MPD). These constitute a heterogeneous group of disorders, also known as myeloproliferative diseases or myeloproliferative neoplasms (MPN), characterized by cellular proliferation of one or more hematologic cell lines in the peripheral blood, distinct from acute leukemia. Included diseases are: essential thrombocythemia, polycythemia vera, primary myelofibrosis (chronic idiopathic myelofibrosis). Bone marrow samples from patients display uniformly low levels of hmC in genomic DNA compared to bone marrow samples from healthy controls as well as hypomethylation relative to controls at the majority of differentially methylated CpG sites. The disease is caused by mutations affecting the gene represented in this entry. TET2 is frequently mutated in systemic mastocytosis; also known as systemic mast cell disease. A condition with features in common with myeloproliferative diseases. It is a clonal disorder of the mast cell and its precursor cells. The clinical symptoms and signs of systemic mastocytosis are due to accumulation of clonally derived mast cells in different tissues, including bone marrow, skin, the gastrointestinal tract, the liver, and the spleen. The disease is caused by mutations affecting the gene represented in this entry. Bone marrow samples from patients display uniformly low levels of hmC in genomic DNA compared to bone marrow samples from healthy controls as well as hypomethylation relative to controls at the majority of differentially methylated CpG sites. FunctionTET2_HUMAN Dioxygenase that catalyzes the conversion of the modified genomic base 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) and plays a key role in active DNA demethylation. Also mediates subsequent conversion of 5hmC into 5-formylcytosine (5fC), and conversion of 5fC to 5-carboxylcytosine (5caC). Conversion of 5mC into 5hmC, 5fC and 5caC probably constitutes the first step in cytosine demethylation. Methylation at the C5 position of cytosine bases is an epigenetic modification of the mammalian genome which plays an important role in transcriptional regulation. In addition to its role in DNA demethylation, also involved in the recruitment of the O-GlcNAc transferase OGT to CpG-rich transcription start sites of active genes, thereby promoting histone H2B GlcNAcylation by OGT.[1] [2] [3] [4] [5] Publication Abstract from PubMedDNA methylation is an important epigenetic modification. Ten-eleven translocation (TET) proteins are involved in DNA demethylation through iteratively oxidizing 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Here we show that human TET1 and TET2 are more active on 5mC-DNA than 5hmC/5fC-DNA substrates. We determine the crystal structures of TET2-5hmC-DNA and TET2-5fC-DNA complexes at 1.80 A and 1.97 A resolution, respectively. The cytosine portion of 5hmC/5fC is specifically recognized by TET2 in a manner similar to that of 5mC in the TET2-5mC-DNA structure, and the pyrimidine base of 5mC/5hmC/5fC adopts an almost identical conformation within the catalytic cavity. However, the hydroxyl group of 5hmC and carbonyl group of 5fC face towards the opposite direction because the hydroxymethyl group of 5hmC and formyl group of 5fC adopt restrained conformations through forming hydrogen bonds with the 1-carboxylate of NOG and N4 exocyclic nitrogen of cytosine, respectively. Biochemical analyses indicate that the substrate preference of TET2 results from the different efficiencies of hydrogen abstraction in TET2-mediated oxidation. The restrained conformation of 5hmC and 5fC within the catalytic cavity may prevent their abstractable hydrogen(s) adopting a favourable orientation for hydrogen abstraction and thus result in low catalytic efficiency. Our studies demonstrate that the substrate preference of TET2 results from the intrinsic value of its substrates at their 5mC derivative groups and suggest that 5hmC is relatively stable and less prone to further oxidation by TET proteins. Therefore, TET proteins are evolutionarily tuned to be less reactive towards 5hmC and facilitate the generation of 5hmC as a potentially stable mark for regulatory functions. Structural insight into substrate preference for TET-mediated oxidation.,Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, Lou Z, Zhang L, Li W, Gong W, Liu M, Sun C, Yin X, Li J, Tan X, Wang P, Wang Y, Fang D, Cui Q, Yang P, He C, Jiang H, Luo C, Xu Y Nature. 2015 Nov 5;527(7576):118-22. doi: 10.1038/nature15713. Epub 2015 Oct 28. PMID:26524525[6] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
| ||||||||||||||||||||
Categories: Homo sapiens | Large Structures | Synthetic construct | Cheng J | Hu L | Li J | Li Z | Rao Q | Xu Y