We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
ACE Inhibitor Prinivil
From Proteopedia
Contents |
Lisinopril
|
[1] was patented by Merck & Co. under the brand name of Prinivil.[2] Lisinopril functions as a competitive inhibitor of the angiotensin converting enzyme (ACE). [3] cleaves specific residues of an inactive near the C domain (domain closer to the C-terminus) to form a potent vasopressor octapeptide known as ; however, the specific substrate binding and catalysis is not fully understood. ACE is found as a type 1 membrane bound dipeptidyl carboxypeptidase that regulates blood pressure and steady state equilibrium of ions within the blood. Located on vascular epithelial cells, the drug interaction takes place on the surface of the cell within an artery/vein.[4] The ligand is stabilized by 3 residues by interacting through hydrogen bonding along with an ionic binding of a atom and the carboxylate group which configures the molecule in 3D space.
Function
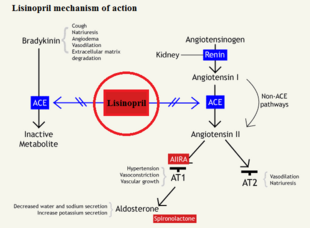
is a common vasodilator that, when bound to ACE, will allow ACE the cleave angiotensin I to angiotensin II, but when bound to lisinopril through competitive inhibition, bradykinin levels rise in the blood stream causing a decrease in blood pressure and increased vasodilation. As seen in Figure 1,[5] bradykinin will bind to the ACE, which allows for the conversion of angiotensin I to angiotensin II by the ACE/bradykinin complex. By blocking the active site of ACE, an inactive angiotensin 1 cannot be cleaved, which would block the cascade of events triggered by angiotensin 2 binding to type 1 AT2 receptor to create vasoconstriction.[6]
Structure and Mechanism
Lisinopril (prinivil) acts upon the membrane protein by forming tight and nonspecific contacts with the conserved residues in the , , and positions of the , which is viewed by using JSmol[7] and Jmol.[8] The S1’ subsite contains Glu162 residue that interacts strongly with the lysine residue of lisinopril, the S1 subsite is surrounded with hydrophobic residues Phe512 and Val518, and the S2’ subsite contains Lys511 and Tyr520 to form strong hydrogen bonds with the C-terminus proline of lisinopril.[9] The ACE Active site has a NH3+ group which binds to the COOH terminus of the drug. NH of the enzyme binds to the middle C=O group of the drug. The benzene ring of the drug fits into a hydrophobic pocket(S1) of the enzyme. The 5-membered nitrogenous ring of the drug fits to the S2’ pocket. The lysine segment fits into the S1’ pocket, as seen in Figure 2.[10] All other ACE inhibitors, such as Captopril, Fentiapril, Pivalopril, etc all have the same general substituents of a phenyl in the S1, lysine in the S1', and proline in the S2' pocket as well as a carboxyl group that binds with a Zn2+ ion.
References
- ↑ Canner, D. Lisinopril http://proteopedia.org/wiki/index.php/prinivil (accessed Nov 10, 2016).
- ↑ PRINIVIL® (lisinopril) https://www.merck.com/product/usa/pi_circulars/p/prinivil/prinivil_pi.pdf (accessed Nov 16, 2016).
- ↑ Akif M, Georgiadis D, Mahajan A, Dive V, Sturrock ED, Isaac RE, Acharya KR. High-resolution crystal structures of Drosophila melanogaster angiotensin-converting enzyme in complex with novel inhibitors and antihypertensive drugs. J Mol Biol. 2010 Jul 16;400(3):502-17. Epub 2010 May 19. PMID:20488190 doi:10.1016/j.jmb.2010.05.024
- ↑ Natesh R, Schwager SL, Sturrock ED, Acharya KR. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003 Jan 30;421(6922):551-4. Epub 2003 Jan 19. PMID:12540854 doi:http://dx.doi.org/10.1038/nature01370
- ↑ Ross, S. ACE Inhibitors. Bpac nc Better Medicine. 2006 http://www.bpac.org.nz/resources/campaign/ace/bpac_ace_poem_2006_wv.pdf
- ↑ Brown, N. Vaughan, D. Angiotensin-Converting Enzyme Inhibitors. Circulation. 1998;97:1411-1420. doi: http://dx.doi.org/10.1161/01.CIR.97.14.1411
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Wang X, Wu S, Xu D, Xie D, Guo H. Inhibitor and substrate binding by angiotensin-converting enzyme: quantum mechanical/molecular mechanical molecular dynamics studies. J Chem Inf Model. 2011 May 23;51(5):1074-82. doi: 10.1021/ci200083f. Epub 2011, Apr 26. PMID:21520937 doi:http://dx.doi.org/10.1021/ci200083f
- ↑ Brew, K. Structure of human ACE gives new insights into inhibitor binding and design. TRENDS in Pharm. Sci. 2003 Aug; 24: 8. doi: http://dx.doi.org/10.1016/S0165-6147(03)00199-8