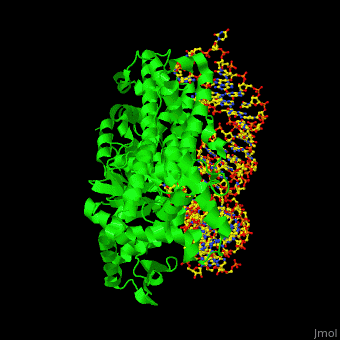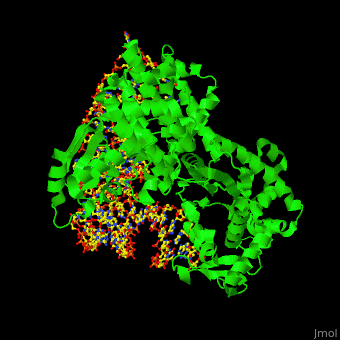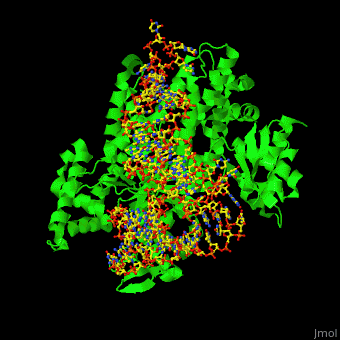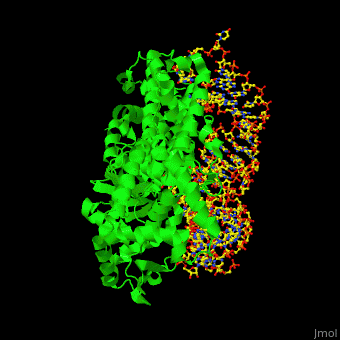
Aminoacyl tRNA synthetase (aaRS) or aminoacyl tRNA ligase catalyzes the esterification of a specific amino acid to its cognate tRNA to form an aminoacyl-tRNA. The amino acid is transferred by the ribosome from the aminoacylated-tRNA onto a growing polypeptide chain. Class I of aaRS is a monomer or dimer, it has 2 highly conserved sequence motifs and it aminoacylates at the 2’-OH of an adenosine nucleotide. Class II of aaRS is a dimer or tetramer, it has 3 highly conserved sequence motifs and it aminoacylates at the 3’-OH of an adenosine nucleotide. CP1 domain of RS edits a mischarged aa-tRNA. Some of the crystal structures are complexes of the RS with their reactant analog: amino acid-sulfamoyl adenine (aa-SA).[1].
See also TRNA synthetases (Hebrew).
3D Structures of Aminoacyl tRNA synthetase
Aminoacyl tRNA synthetase 3D structures
One very interesting question in biology is how does an aminoacyl-tRNA synthetase recognize a particular tRNA and charge it with the correct amino acid? This is a challenging problem, since all tRNAs have the same general structure. Interestingly, different tRNA synthetases accomplish this goal in different ways.
The (GlnRS) interacts with both . Genetic and biochemical data indicate that GlnRS interacts with all three bases of the , which are unstacked and splay outward so they can bind in separate recognition pockets of GlnRS. The 3' end of tRNAgln plunges deeply into a protein pocket that also binds the enzyme's and glutamine substrates.
One interesting question is how tRNA synthetases are able to discern that the correct amino acid is loaded. The structure for aspartate is very similar to glutamine, differing only by an amino group and one CH2 group. However, the aminoacyl tRNA synthetases interact with the tRNA in completely different fashions. Asp-tRNA synthetase acts as a , and interacts with the opposite face of the tRNA, the side. The anticodon arm of tRNA Asp is bent by as much as 20 A toward the inside of the L relative to that in the X-ray structure of uncomplexed tRNA Asp, and its anticodon bases are unstacked. The hinge point for the bend is a base pair in the anticodon stem (nearly all other species of tRNA contain a Watson-Crick base pair at that point)