Azoarcus group I intron
From Proteopedia
| For the date when the most recent work on this article was done, click on the history tab above. |
|
CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS
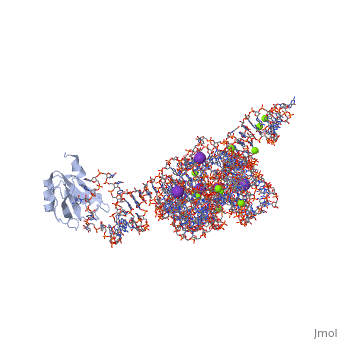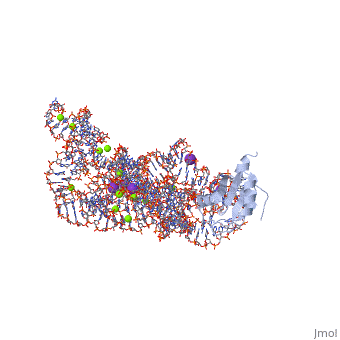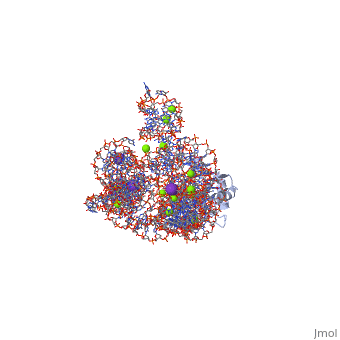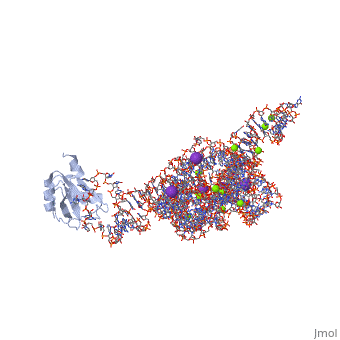
The discovery of the RNA self-splicing group I intron provided the first demonstration that not all enzymes are proteins. Here we report the X-ray crystal structure (3.1-A resolution) of a complete group I bacterial intron in complex with both the 5'- and the 3'-exons. This complex corresponds to the splicing intermediate before the exon ligation step. It reveals how the intron uses structurally unprecedented RNA motifs to select the 5'- and 3'-splice sites. The 5'-exon's 3'-OH is positioned for inline nucleophilic attack on the conformationally constrained scissile phosphate at the intron-3'-exon junction. Six phosphates from three disparate RNA strands converge to coordinate two metal ions that are asymmetrically positioned on opposing sides of the reactive phosphate. This structure represents the first splicing complex to include a complete intron, both exons and an organized active site occupied with metal ions.
Crystal structure of a self-splicing group I intron with both exons., Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA, Nature. 2004 Jul 1;430(6995):45-50. Epub 2004 Jun 2. PMID:15175762
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
1U6B is a 4 chains structure with sequences from Azoarcus. This structure supersedes the now removed PDB entry 1t42. The May 2005 RCSB PDB Molecule of the Month feature on Self-splicing RNA by David S. Goodsell is 10.2210/rcsb_pdb/mom_2005_5. Full crystallographic information is available from OCA.
Reference
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004 Jul 1;430(6995):45-50. Epub 2004 Jun 2. PMID:15175762 doi:10.1038/nature02642
Bold text