SEE ALSO CRISPR-Cas
I-F CRISPR/Cas system of Pseudomonas aeruginosa
Anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with a 32nt spacer crRNA backbone region[1]
Bacteriophages encode anti-CRISPR suppressors to counteract the CRISPR/Cas immunity of their bacterial hosts, thus facilitating their survival and replication. Previous studies have shown that two phage-encoded anti-CRISPR proteins, AcrF1 and AcrF2, suppress the type I-F CRISPR/Cas system of Pseudomonas aeruginosa by preventing target DNA recognition by the Csy surveillance complex, but the precise underlying mechanism was unknown. Here the structure of AcrF1/2 bound to the Csy complex determined by cryo-EM single-particle reconstruction was presented. By structural analysis, was found that AcrF1 inhibits target DNA recognition of the Csy complex by interfering with base pairing between the DNA target strand and crRNA spacer. In addition, multiple copies of AcrF1 bind to the Csy complex with different modes when working individually or cooperating with AcrF2, which might exclude target DNA binding through different mechanisms. Together with previous reports, a comprehensive working scenario was provided for the two anti-CRISPR suppressors, AcrF1 and AcrF2, which silence CRISPR/Cas immunity by targeting the Csy surveillance complex.
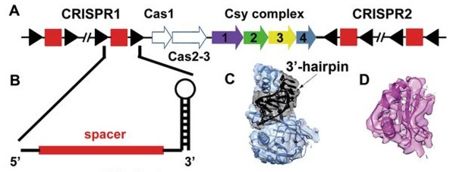
Fig. 1 (A) Schematic representation of the CRISPR/Cas gene clusters of P. aeruginosa. The CRISPR repeats and spacers are represented as black triangles and red squares, respectively. The Cas2 protein in this system is fused as a domain in the Cas3 protein. The four Csy proteins consisting of the Csy complex (Csy1-4) are consecutively arranged in the genome and colored in purple, green, yellow and slate, respectively. (B) Close view of the schematic model of a mature crRNA with the spacer colored in red and repeat sequence in black. (C) The density of Csy4-crRNA hairpin head and the fitted atomic model. The Csy4 protein is represented by slate ribbons and the crRNA 3′-hairpin is highlighted in black. (D) Representative density of AcrF1 and the refined atomic model. The model is shown as ribbons and the density map shows clear side chain features. From
[1]Anti-CRISPR proteins AcrF1/2 bound to Csy surveillance complex with a 20nt spacer crRNA backbone region
Additional representatives of CRISPR type I-F
- Pseudomonas aeruginosa: 2xlj, 2xlk, 4al5, 4al6, 4al7, 5uz9, 6b46, 6b48, 6b47, 6b45, 6b44, 6anw, 6anv, 5gqh, 5gnf.
- Shewanella putrefaciens: 5o6u, 5o7h

