We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Forkhead Box Protein 3
From Proteopedia
(Redirected from FOXP3)
| |||||||||||
References
- ↑ Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009 May;30(5):616-25. PMID:19464984 doi:10.1016/j.immuni.2009.04.009
- ↑ Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007 Feb 22;445(7130):936-40. Epub 2007 Jan 21. PMID:17237761 doi:10.1038/nature05563
- ↑ Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009 Nov;10(11):1170-7. Epub 2009 Sep 20. PMID:19767756 doi:10.1038/ni.1795
- ↑ Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001 Jan;27(1):20-1. PMID:11137993 doi:10.1038/83713
- ↑ Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007 Mar;8(3):277-84. Epub 2007 Jan 14. PMID:17220892 doi:10.1038/ni1437
- ↑ Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003 Apr;4(4):330-6. Epub 2003 Mar 3. PMID:12612578 doi:10.1038/ni904
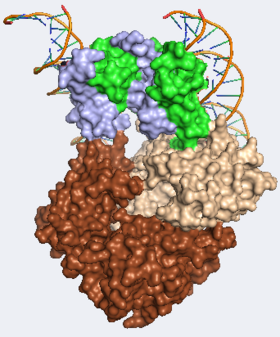
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Bandukwala HS, Wu Y, Feurer M, Chen Y, Barbosa B, Ghosh S, Stroud JC, Benoist C, Mathis D, Rao A, Chen L. Structure of a Domain-Swapped FOXP3 Dimer on DNA and Its Function in Regulatory T Cells. Immunity. 2011 Mar 30. PMID:21458306 doi:10.1016/j.immuni.2011.02.017
- ↑ Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, Azzari C, Ziegler SF, Levings MK, Roncarolo MG. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J Clin Invest. 2006 Jun;116(6):1713-22. PMID:16741580 doi:10.1172/JCI25112