Grb10 SH2 Domain
From Proteopedia
| |||||||||||
Contents |
Interaction Between Grb10 and E3 Ubiquitin Ligase NEDD4
|
Grb10 has now been shown to not only inhibit insulin receptors and IGF1R kinase activity, but also interacts via its SH2 domain with the C2 domain of E3 ubiquitin ligase NEDD4 facilitating ubiquitation of IGF1R [3]. It is hypothesized that the Grb10 SH2 interaction with the E3 domain of NEDD4 may allow NEDD4 to come in close proximity to IGF1R promoting degradation.[4]
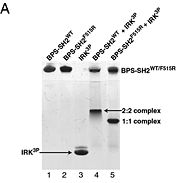
There are 3 interfaces at which Nedd4 C2 and Grb10 SH2 interact:
is the largest of the 3 interfaces between Grb10 SH2 and NEDD4 C2 domains. There are 8 hydrogen bonds formed at this interface ( and ), with the least important H bonds formed between , for disruption between these two residues with an induced mutation did not inhibit Grb10 SH2 and NEDD4 C2 to complex. [5]
, the smallest of the 3, is composed of residues 429-434 of Grb10 SH2 interacting with residues 112-114 of NEDD4 C2; disruption of this interaction does not disrupt the SH2-C2 domain complex. [6]
is made of NEDD4 C2's proline rich C terminus which complexes with Grb10 H2 N-terminal residues 431-440 and C-terminal residues 532-535; among this interaction forms a hydrogen bond and forms a slat bridge. [7]
Grb10 Gene Inhibition Affects Body Composition, and Insulin Signaling
Many studies have shown that when mice are subjected to Grb10 gene disruption (chromosome 11) during prenatal life, they become approximately 30% larger in muscle mass, have improved glucose homeostasis, improved insulin sensitivity and reduced adiposity, i.e. fat storage, during their postnatal life [8] [9].
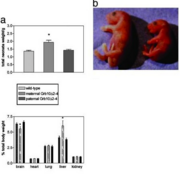
Picture 2 shows how inhibition of the Grb10 affects the overall growth of a mouse fetus. [10] In section a, it is shown that the wild-type and paternal Grb10 knockout mice have virtually the same weight, suggesting that it is not the paternal Grb10 allele that inhibits overally body mass. The middle bar represents the maternal knockout Grb10 mouse and shows an approximate 30% increase in overall weight when compare to wild-type. This data is visualized in section b, where the maternal Grb10 knockout mouse (left) sows a significant size difference when compared tot he wild-type mouse (right). Directly below section a, a bar graph representing the percent total body weight of specific vital organs. The liver shows an overall increase in percent body mass while the brain shows a slight decrease. More research must be done to determine why the brain and liver are specifically affected by Grb10 inhibition.
Medical Implications for Grb10 Inhibition
The Grb10 gene is not only found in mice but also in humans on chromosome 7p11.2–p12. Approximately 10% of patients whom suffer from Russell-Silver Syndrome, a disease associated with severe growth retardation, have a defect in chromosome 7, suggesting a linkage between Grb10 and poor growth in humans.[11]
In recent news, Grb10 has been dubbed the "hulk protein" due to new findings of a new study from the Garvan Institute of Medical Research in Sydney, Australia. Researchers sought to find the skeletal muscle physiology of mice that had Grb10 deleted from their genome. They found that the enlargement of muscles was due to the increase in numbers of myofibers in the muscle tissue, not myofiber size. [12] The investigators also found that the Grb10-deficient mice muscles were metabolically no different then muscles found in wild-type mice. [13]
Once these findings were published, the media had a hay day writing about the newly discovered "Hulk" Protein. Here are the titles of a few news articles found online regarding Grb10:
"Scientists Discover 'Hulk' Protein"
"Newly Discovered 'Hulk' Protein Could Make You a Beefcake Without the Weights"
"Growing Strong Muscles Without Working Out? 'Hulk' Protein, Grb10, Controls Muscle Growth"
Dr. Lowenna J. Holt, Ph.D., the lead researcher was quoted saying, "By identifying a novel mechanism regulating muscle development, our work has revealed potential new strategies to increase muscle mass. Ultimately, this might improve treatment of muscle wasting conditions, as well as metabolic disorders such as Type 2 diabetes". Dr. Holt and colleagues did not set out to find a new way to increase the muscle mass for gym rats or competitive athletes but rather a new approach to treat patients that suffer from many different muscle wasting diseases. A lot more research must be done before it can be used on humans, but we may one day see the olympics testing for Grb10 inhibitors for they will most likely be abused as a new modern alternative to steroids.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Stein EG, Ghirlando R, Hubbard SR. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J Biol Chem. 2003 Apr 11;278(15):13257-64. Epub 2003 Jan 27. PMID:12551896 doi:http://dx.doi.org/10.1074/jbc.M212026200
- ↑ He W, Rose DW, Olefsky JM, Gustafson TA. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem. 1998 Mar 20;273(12):6860-7. PMID:9506989
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, Cooney GJ, Daly RJ, Ward A. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 2007 Aug;27(16):5871-86. Epub 2007 Jun 11. PMID:17562854 doi:10.1128/MCB.02087-06
- ↑ Huang Q, Szebenyi DM. Structural basis for the interaction between the growth factor-binding protein GRB10 and the E3 ubiquitin ligase NEDD4. J Biol Chem. 2010 Dec 31;285(53):42130-9. Epub 2010 Oct 26. PMID:20980250 doi:10.1074/jbc.M110.143412
- ↑ Morita H, Omichi M, Inami I, Koike S. [Role of hepatic cycle AMP and lysosomal lipase in triglyceridemia induced by injection of cobalt chloride]. Nihon Eiseigaku Zasshi. 1975 Apr;30(1):65. PMID:166222
- ↑ Smith FM, Holt LJ, Garfield AS, Charalambous M, Koumanov F, Perry M, Bazzani R, Sheardown SA, Hegarty BD, Lyons RJ, Cooney GJ, Daly RJ, Ward A. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 2007 Aug;27(16):5871-86. Epub 2007 Jun 11. PMID:17562854 doi:10.1128/MCB.02087-06
- ↑ Holt LJ, Turner N, Mokbel N, Trefely S, Kanzleiter T, Kaplan W, Ormandy CJ, Daly RJ, Cooney GJ. Grb10 regulates the development of fiber number in skeletal muscle. FASEB J. 2012 Sep;26(9):3658-69. doi: 10.1096/fj.11-199349. Epub 2012 May 23. PMID:22623587 doi:10.1096/fj.11-199349
- ↑ Name= Lowenna PMID: 22623587